Inter- and intra-reproducibility of genotypes from sheep technical replicates on Illumina and Affymetrix platforms
- PMID: 27832740
- PMCID: PMC5105264
- DOI: 10.1186/s12711-016-0267-0
Inter- and intra-reproducibility of genotypes from sheep technical replicates on Illumina and Affymetrix platforms
Abstract
Background: Accurate genomic analyses are predicated upon access to accurate genotype input data. The objective of this study was to quantify the reproducibility of genotype data that are generated from the same genotype platform and from different genotyping platforms.
Methods: Genotypes based on 51,121 single nucleotide polymorphisms (SNPs) for 84 animals that were each genotyped on Illumina and Affymetrix platforms and for another 25 animals that were each genotyped twice on the same Illumina platform were compared. Genotypes based on 11,323 SNPs for an additional 21 animals that were genotyped on two different Illumina platforms by two different service providers were also compared. Reproducibility of the results was measured as the correlation between allele counts and as genotype and allele concordance rates.
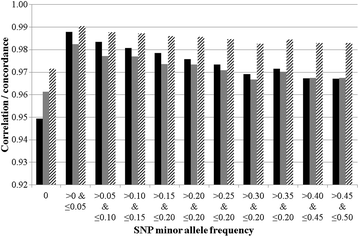
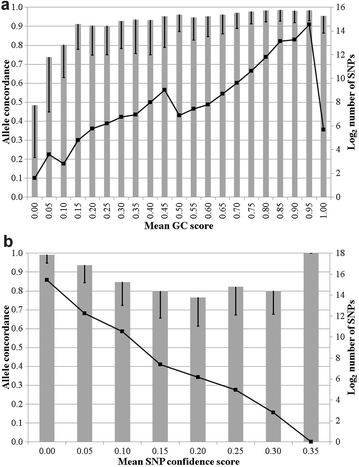
Results: A mean within-animal correlation of 0.9996 was found between allele counts in the 25 duplicate samples that were genotyped on the same Illumina platform and varied from 0.9963 to 1.0000 per animal. The mean (minimum, maximum) genotype and allele concordance rates per animal between the 25 duplicate samples were equal to 0.9996 (0.9968, 1.0000) and 0.9993 (0.9937, 1.0000), respectively. The concordance rate between the two different Illumina platforms was also near 1. A mean within-animal correlation of 0.9738 was found between genotypes that were generated on the Illumina and Affymetrix platforms and varied from 0.9505 to 0.9812 per animal. The mean (minimum, maximum) within-animal genotype and allele concordance rates between the Illumina and Affymetrix platforms were equal to 0.9711 (0.9418, 0.9798) and 0.9845 (0.9695, 0.9889), respectively. The genotype concordance rate across all genotypes increased from 0.9711 to 0.9949 when the SNPs used were restricted to those with three high-resolution genotype clusters which represented 75.2% of the called genotypes.
Conclusions and implications: Our results suggest that, regardless of the genotype platform or service provider, high genotype concordance rates are achieved especially if they are restricted to high-quality extracted DNA and SNPs that result in high-quality genotypes.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

