Genetic tools for advancement of Synechococcus sp. PCC 7002 as a cyanobacterial chassis
- PMID: 27832791
- PMCID: PMC5105302
- DOI: 10.1186/s12934-016-0584-6
Genetic tools for advancement of Synechococcus sp. PCC 7002 as a cyanobacterial chassis
Abstract
Background: Successful implementation of modified cyanobacteria as hosts for industrial applications requires the development of a cyanobacterial chassis. The cyanobacterium Synechococcus sp. PCC 7002 embodies key attributes for an industrial host, including a fast growth rate and high salt, light, and temperature tolerances. This study addresses key limitations in the advancement of Synechococcus sp. PCC 7002 as an industrial chassis.
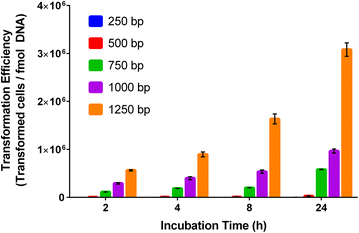
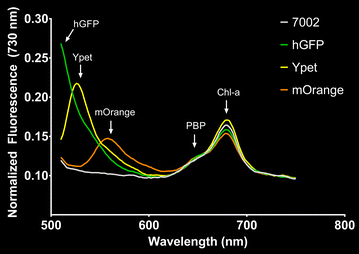
Results: Tools for genome integration were developed and characterized, including several putative neutral sites for genome integration. The minimum homology arm length for genome integration in Synechococcus sp. PCC 7002 was determined to be approximately 250 bp. Three fluorescent protein reporters (hGFP, Ypet, and mOrange) were characterized for gene expression, microscopy, and flow cytometry applications in Synechococcus sp. PCC 7002. Of these three proteins, the yellow fluorescent protein (Ypet) had the best optical properties for minimal interference with the native photosynthetic pigments and for detection using standard microscopy and flow cytometry optics. Twenty-five native promoters were characterized as tools for recombinant gene expression in Synechococcus sp. PCC 7002 based on previous RNA-seq results. This characterization included comparisons of protein and mRNA levels as well as expression under both continuous and diurnal light conditions. Promoters A2520 and A2579 were found to have strong expression in Synechococcus sp. PCC 7002 while promoters A1930, A1961, A2531, and A2813 had moderate expression. Promoters A2520 and A2813 showed more than twofold increases in gene expression under light conditions compared to dark, suggesting these promoters may be useful tools for engineering diurnal regulation.
Conclusions: The genome integration, fluorescent protein, and promoter tools developed in this study will help to advance Synechococcus sp. PCC 7002 as a cyanobacterial chassis. The long minimum homology arm length for Synechococcus sp. PCC 7002 genome integration indicates native exonuclease activity or a low efficiency of homologous recombination. Low correlation between transcript and protein levels in Synechococcus sp. PCC 7002 suggests that transcriptomic data are poor selection criteria for promoter tool development. Lastly, the conventional strategy of using promoters from photosynthetic operons as strong promoter tools is debunked, as promoters from hypothetical proteins (A2520 and A2579) were found to have much higher expression levels.
Keywords: Cyanobacterial cell factories; Cyanobacterial chassis; Cyanobacterial genetic engineering; Cyanobacterial host; Synechococcus; Synechococcus 7002; Synechococcus sp. PCC 7002.
Figures

Similar articles
-
Genetic, Genomics, and Responses to Stresses in Cyanobacteria: Biotechnological Implications.Genes (Basel). 2021 Mar 29;12(4):500. doi: 10.3390/genes12040500. Genes (Basel). 2021. PMID: 33805386 Free PMC article. Review.
-
Zn2+-Inducible Expression Platform for Synechococcus sp. Strain PCC 7002 Based on the smtA Promoter/Operator and smtB Repressor.Appl Environ Microbiol. 2017 Jan 17;83(3):e02491-16. doi: 10.1128/AEM.02491-16. Print 2017 Feb 1. Appl Environ Microbiol. 2017. PMID: 27836841 Free PMC article.
-
A toolbox to engineer the highly productive cyanobacterium Synechococcus sp. PCC 11901.Plant Physiol. 2024 Oct 1;196(2):1674-1690. doi: 10.1093/plphys/kiae261. Plant Physiol. 2024. PMID: 38713768 Free PMC article.
-
Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973.Metab Eng. 2018 Jul;48:163-174. doi: 10.1016/j.ymben.2018.06.002. Epub 2018 Jun 6. Metab Eng. 2018. PMID: 29883802
-
Fast-growing cyanobacterial chassis for synthetic biology application.Crit Rev Biotechnol. 2024 May;44(3):414-428. doi: 10.1080/07388551.2023.2166455. Epub 2023 Feb 26. Crit Rev Biotechnol. 2024. PMID: 36842999 Review.
Cited by
-
Synthetic biology in marine cyanobacteria: Advances and challenges.Front Microbiol. 2022 Sep 16;13:994365. doi: 10.3389/fmicb.2022.994365. eCollection 2022. Front Microbiol. 2022. PMID: 36188008 Free PMC article. Review.
-
The Synthetic Biology Toolkit for Photosynthetic Microorganisms.Plant Physiol. 2019 Sep;181(1):14-27. doi: 10.1104/pp.19.00345. Epub 2019 Jul 1. Plant Physiol. 2019. PMID: 31262955 Free PMC article. Review.
-
Proximity-based proteomics reveals the thylakoid lumen proteome in the cyanobacterium Synechococcus sp. PCC 7002.Photosynth Res. 2021 Feb;147(2):177-195. doi: 10.1007/s11120-020-00806-y. Epub 2020 Dec 6. Photosynth Res. 2021. PMID: 33280076 Free PMC article.
-
Application of Cyanobacteria as Chassis Cells in Synthetic Biology.Microorganisms. 2024 Jul 5;12(7):1375. doi: 10.3390/microorganisms12071375. Microorganisms. 2024. PMID: 39065143 Free PMC article. Review.
-
Genetic, Genomics, and Responses to Stresses in Cyanobacteria: Biotechnological Implications.Genes (Basel). 2021 Mar 29;12(4):500. doi: 10.3390/genes12040500. Genes (Basel). 2021. PMID: 33805386 Free PMC article. Review.
References
-
- Dexter J, Fu PC. Metabolic engineering of cyanobacteria for ethanol production. Energy Environ Sci. 2009;2:857. doi: 10.1039/b811937f. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

