Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits
- PMID: 27833659
- PMCID: PMC5103450
- DOI: 10.1186/s13072-016-0102-4
Identification of activated enhancers and linked transcription factors in breast, prostate, and kidney tumors by tracing enhancer networks using epigenetic traits
Abstract
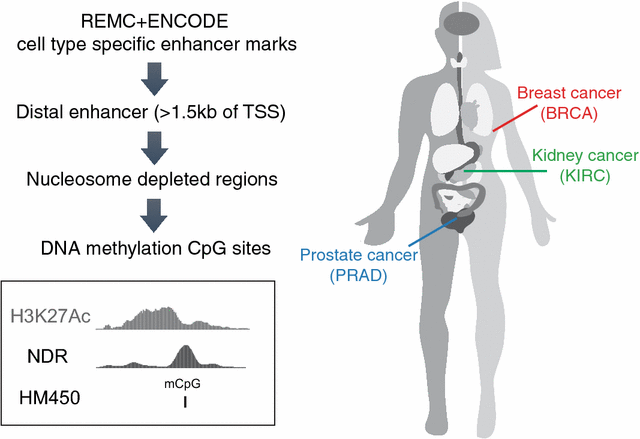
Background: Although technological advances now allow increased tumor profiling, a detailed understanding of the mechanisms leading to the development of different cancers remains elusive. Our approach toward understanding the molecular events that lead to cancer is to characterize changes in transcriptional regulatory networks between normal and tumor tissue. Because enhancer activity is thought to be critical in regulating cell fate decisions, we have focused our studies on distal regulatory elements and transcription factors that bind to these elements.
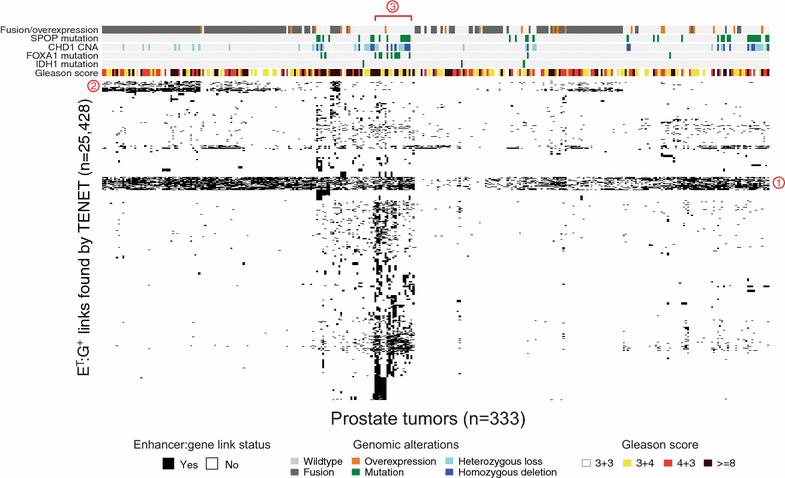
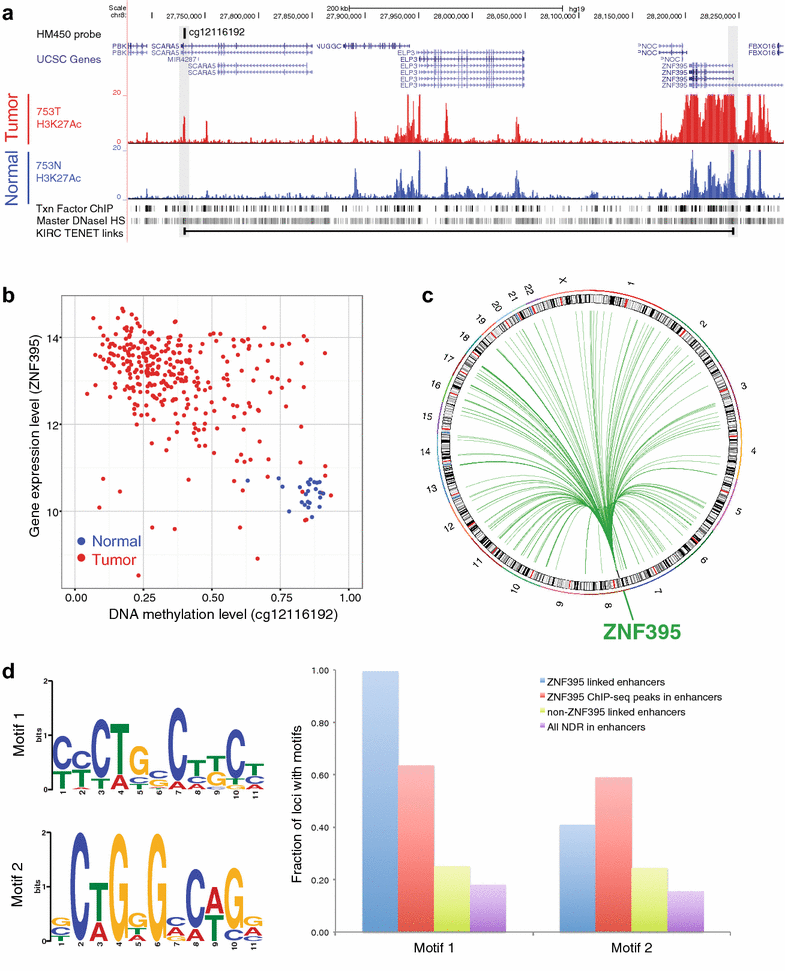
Results: Using DNA methylation data, we identified more than 25,000 enhancers that are differentially activated in breast, prostate, and kidney tumor tissues, as compared to normal tissues. We then developed an analytical approach called Tracing Enhancer Networks using Epigenetic Traits that correlates DNA methylation levels at enhancers with gene expression to identify more than 800,000 genome-wide links from enhancers to genes and from genes to enhancers. We found more than 1200 transcription factors to be involved in these tumor-specific enhancer networks. We further characterized several transcription factors linked to a large number of enhancers in each tumor type, including GATA3 in non-basal breast tumors, HOXC6 and DLX1 in prostate tumors, and ZNF395 in kidney tumors. We showed that HOXC6 and DLX1 are associated with different clusters of prostate tumor-specific enhancers and confer distinct transcriptomic changes upon knockdown in C42B prostate cancer cells. We also discovered de novo motifs enriched in enhancers linked to ZNF395 in kidney tumors.
Conclusions: Our studies characterized tumor-specific enhancers and revealed key transcription factors involved in enhancer networks for specific tumor types and subgroups. Our findings, which include a large set of identified enhancers and transcription factors linked to those enhancers in breast, prostate, and kidney cancers, will facilitate understanding of enhancer networks and mechanisms leading to the development of these cancers.
Keywords: DNA methylation; Enhancer; Epigenetics; Networks; Transcription factor.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

