ZHX1 Promotes the Proliferation, Migration and Invasion of Cholangiocarcinoma Cells
- PMID: 27835650
- PMCID: PMC5105949
- DOI: 10.1371/journal.pone.0165516
ZHX1 Promotes the Proliferation, Migration and Invasion of Cholangiocarcinoma Cells
Abstract
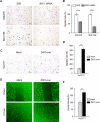
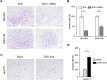
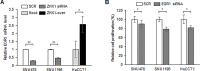
Zinc-fingers and homeoboxes 1 (ZHX1) is a transcription repressor that has been associated with the progressions of hepatocellular carcinoma, gastric cancer, and breast cancer. However, the functional roles of ZHX1 in cholangiocarcinoma (CCA) have not been determined. We investigated the expression and roles of ZHX1 during the proliferation, migration, and invasion of CCA cells. In silico analysis and immunohistochemical studies showed amplification and overexpression of ZHX1 in CCA tissues. Furthermore, ZHX1 knockdown using specific siRNAs decreased CCA cell proliferation, migration, and invasion, whereas ZHX1 overexpression promoted all three characteristics. In addition, results suggested EGR1 might partially mediate the effect of ZHX1 on the proliferation of CCA cells. Taken together, these results show ZHX1 promotes CCA cell proliferation, migration, and invasion, and present ZHX1 as a potential target for the treatment of CCA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells.Biochem Biophys Res Commun. 2018 Feb 5;496(2):455-461. doi: 10.1016/j.bbrc.2018.01.077. Epub 2018 Jan 12. Biochem Biophys Res Commun. 2018. PMID: 29337065
-
Roles of zinc-fingers and homeoboxes 1 during the proliferation, migration, and invasion of glioblastoma cells.Tumour Biol. 2017 Mar;39(3):1010428317694575. doi: 10.1177/1010428317694575. Tumour Biol. 2017. PMID: 28351300
-
MicroRNA-144 suppresses cholangiocarcinoma cell proliferation and invasion through targeting platelet activating factor acetylhydrolase isoform 1b.BMC Cancer. 2014 Dec 5;14:917. doi: 10.1186/1471-2407-14-917. BMC Cancer. 2014. PMID: 25479763 Free PMC article.
-
Molecular pathogenesis of cholangiocarcinoma.Dig Dis. 2014;32(5):564-9. doi: 10.1159/000360502. Epub 2014 Jul 14. Dig Dis. 2014. PMID: 25034289 Free PMC article. Review.
-
Functions and roles of long noncoding RNA in cholangiocarcinoma.J Cell Physiol. 2019 Aug;234(10):17113-17126. doi: 10.1002/jcp.28470. Epub 2019 Mar 19. J Cell Physiol. 2019. PMID: 30888066 Review.
Cited by
-
Cooperative regulation of Zhx1 and hnRNPA1 drives the cardiac progenitor-specific transcriptional activation during cardiomyocyte differentiation.Cell Death Discov. 2023 Jul 14;9(1):244. doi: 10.1038/s41420-023-01548-1. Cell Death Discov. 2023. PMID: 37452012 Free PMC article.
-
Attenuated ZHX3 expression is predictive of poor outcome for liver cancer: Indication for personalized therapy.Oncol Lett. 2022 May 25;24(1):224. doi: 10.3892/ol.2022.13345. eCollection 2022 Jul. Oncol Lett. 2022. PMID: 35720472 Free PMC article.
-
Decreased zinc-fingers and homeoboxes family expression was associated with unfavorable outcomes and immune infiltration in lung adenocarcinoma.Transl Cancer Res. 2023 Jun 30;12(6):1422-1440. doi: 10.21037/tcr-22-2793. Epub 2023 Jun 7. Transl Cancer Res. 2023. PMID: 37434685 Free PMC article.
-
Attenuated ZHX3 expression serves as a potential biomarker that predicts poor clinical outcomes in breast cancer patients.Cancer Manag Res. 2019 Feb 5;11:1199-1210. doi: 10.2147/CMAR.S184340. eCollection 2019. Cancer Manag Res. 2019. PMID: 30787639 Free PMC article.
-
Shank-associated RH domain-interacting protein expression is upregulated in entodermal and mesodermal cancer or downregulated in ectodermal malignancy.Oncol Lett. 2018 Dec;16(6):7180-7188. doi: 10.3892/ol.2018.9514. Epub 2018 Sep 27. Oncol Lett. 2018. PMID: 30546455 Free PMC article.
References
-
- Segura-Lopez FK, Aviles-Jimenez F, Guitron-Cantu A, Valdez-Salazar HA, Leon-Carballo S, Guerrero-Perez L, et al. Infection with Helicobacter bilis but not Helicobacter hepaticus was Associated with Extrahepatic Cholangiocarcinoma. Helicobacter. 2015;20(3):223–30. Epub 2015/01/15. 10.1111/hel.12195 . - DOI - PubMed
-
- de Jong MC, Nathan H, Sotiropoulos GC, Paul A, Alexandrescu S, Marques H, et al. Intrahepatic cholangiocarcinoma: an international multi-institutional analysis of prognostic factors and lymph node assessment. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(23):3140–5. Epub 2011/07/07. 10.1200/jco.2011.35.6519 . - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

