Enhancement of 5-FU sensitivity by the proapoptotic rpL3 gene in p53 null colon cancer cells through combined polymer nanoparticles
- PMID: 27835895
- PMCID: PMC5346744
- DOI: 10.18632/oncotarget.13216
Enhancement of 5-FU sensitivity by the proapoptotic rpL3 gene in p53 null colon cancer cells through combined polymer nanoparticles
Abstract
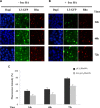
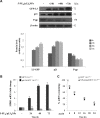
Colon cancer is one of the leading causes of cancer-related death worldwide and the therapy with 5-fluorouracil (5-FU) is mainly limited due to resistance. Recently, we have demonstrated that nucleolar stress upon 5-FU treatment leads to the activation of ribosome-free rpL3 (L3) as proapoptotic factor. In this study, we analyzed L3 expression profile in colon cancer tissues and demonstrated that L3 mRNA amount decreased with malignant progression and the intensity of its expression was inversely related to tumor grade and Bcl-2/Bax ratio. With the aim to develop a combined therapy of 5-FU plus plasmid encoding L3 (pL3), we firstly assessed the potentiation of the cytotoxic effect of 5-FU on colon cancer cells by L3. Next, 10 μM 5-FU and 2 μg of pL3 were encapsulated in biocompatible nanoparticles (NPs) chemically conjugated with HA to achieve active tumor-targeting ability in CD44 overexpressing cancer cells. We showed the specific intracellular accumulation of NPs in cells and a sustained release for 5-FU and L3. Analysis of cytotoxicity and apoptotic induction potential of combined NPs clearly showed that the 5-FU plus L3 were more effective in inducing apoptosis than 5-FU or L3 alone. Furthermore, we show that the cancer-specific chemosensitizer effect of combined NPs may be dependent on L3 ability to affect 5-FU efflux by controlling P-gp (P-glycoprotein) expression. These results led us to propose a novel combined therapy with the use of 5-FU plus L3 in order to establish individualized therapy by examining L3 profiles in tumors to yield a better clinical outcomes.
Keywords: 5-FU; apoptosis; colon cancer; p53; ribosomal protein rpL3.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

