Hypoxia response in Arabidopsis roots infected by Plasmodiophora brassicae supports the development of clubroot
- PMID: 27835985
- PMCID: PMC5106811
- DOI: 10.1186/s12870-016-0941-y
Hypoxia response in Arabidopsis roots infected by Plasmodiophora brassicae supports the development of clubroot
Abstract
Background: The induction of alcohol fermentation in roots is a plant adaptive response to flooding stress and oxygen deprivation. Available transcriptomic data suggest that fermentation-related genes are also frequently induced in roots infected with gall forming pathogens, but the biological significance of this induction is unclear. In this study, we addressed the role of hypoxia responses in Arabidopsis roots during infection by the clubroot agent Plasmodiophora brassicae.
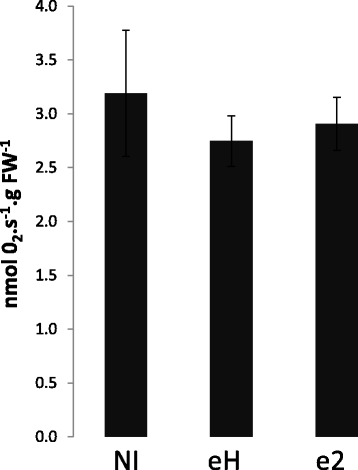
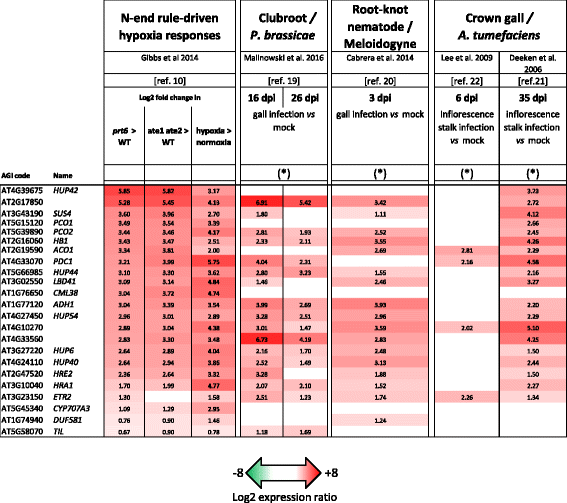
Results: The hypoxia-related gene markers PYRUVATE DECARBOXYLASE 1 (PDC1), PYRUVATE DECARBOXYLASE 2 (PDC2) and ALCOHOL DEHYDROGENASE 1 (ADH1) were induced during secondary infection by two isolates of P. brassicae, eH and e2. PDC2 was highly induced as soon as 7 days post inoculation (dpi), i.e., before the development of gall symptoms, and GUS staining revealed that ADH1 induction was localised in infected cortical cells of root galls at 21 dpi. Clubroot symptoms were significantly milder in the pdc1 and pdc2 mutants compared with Col-0, but a null T-DNA insertional mutation of ADH1 did not affect clubroot susceptibility. The Arg/N-end rule pathway of ubiquitin-mediated proteolysis controls oxygen sensing in plants. Mutants of components of this pathway, ate1 ate2 and prt6, that both exhibit constitutive hypoxia responses, showed enhanced clubroot symptoms. In contrast, gall development was reduced in quintuple and sextuple mutants where the activity of all oxygen-sensing Group VII Ethylene Response Factor transcription factors (ERFVIIs) is absent (erfVII and prt6 erfVII).
Conclusions: Our data demonstrate that the induction of PDC1 and PDC2 during the secondary infection of roots by P. brassicae contributes positively to clubroot development, and that this is controlled by oxygen-sensing through ERFVIIs. The absence of any major role of ADH1 in symptom development may also suggest that PDC activity could contribute to the formation of galls through the activation of a PDH bypass.
Keywords: ADH1; Arabidopsis; Clubroot; ERFVII; Ethanol fermentation; Hypoxia; N-end rule pathway; PDC2; Plant gall disease; Plasmodiophora.
Figures




References
-
- Kageyama K, Asano T. Life Cycle of Plasmodiophora brassicae. J Plant Growth Regul. 2009;28:203–211. doi: 10.1007/s00344-009-9101-z. - DOI
-
- Ludwig-Muller J, Prinsen E, Rolfe SA, Scholes JD. Metabolism and Plant Hormone Action During Clubroot Disease. J Plant Growth Regul. 2009;28:229–244. doi: 10.1007/s00344-009-9089-4. - DOI
-
- Jubault M, Lariagon C, Taconnat L, Renou J-P, Gravot A, Delourme R, et al. Partial resistance to clubroot in Arabidopsis is based on changes in the host primary metabolism and targeted cell division and expansion capacity. Funct Integr Genomics. 2013;13:191–205. doi: 10.1007/s10142-013-0312-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

