The evolving world of pseudoenzymes: proteins, prejudice and zombies
- PMID: 27835992
- PMCID: PMC5106787
- DOI: 10.1186/s12915-016-0322-x
The evolving world of pseudoenzymes: proteins, prejudice and zombies
Abstract
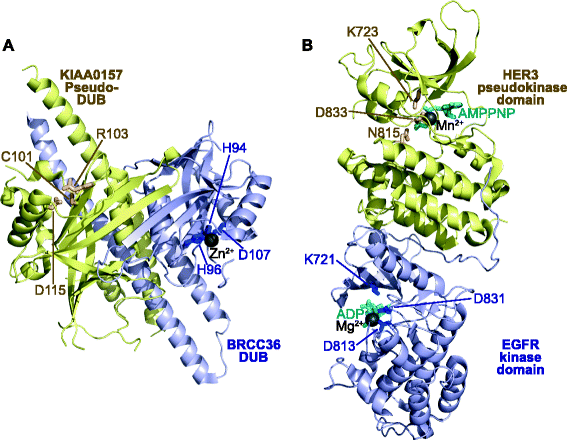
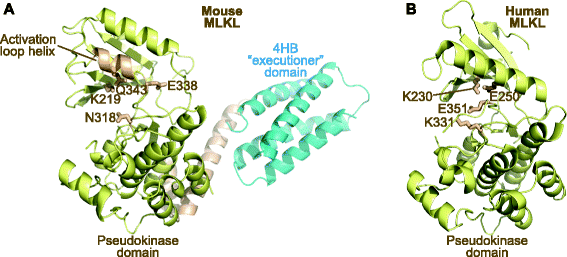
Pseudoenzymes are catalytically deficient variants of enzymes that are represented in all major enzyme families. Their regulatory functions in signalling pathways are shedding new light on the non-catalytic functions of active enzymes, and are suggesting new ways to target cellular signalling mechanisms with drugs.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

