Novel genetic risk variants for pediatric celiac disease
- PMID: 27836013
- PMCID: PMC5105295
- DOI: 10.1186/s40246-016-0091-1
Novel genetic risk variants for pediatric celiac disease
Abstract
Background: Celiac disease is a complex chronic immune-mediated disorder of the small intestine. Today, the pathobiology of the disease is unclear, perplexing differential diagnosis, patient stratification, and decision-making in the clinic.
Methods: Herein, we adopted a next-generation sequencing approach in a celiac disease trio of Greek descent to identify all genomic variants with the potential of celiac disease predisposition.
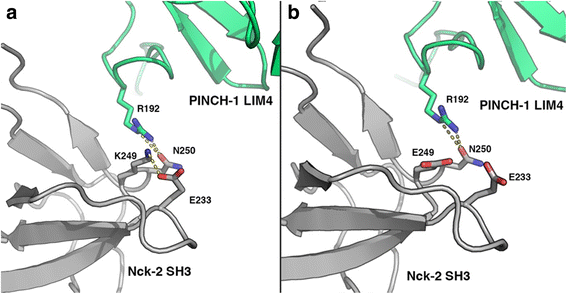
Results: Analysis revealed six genomic variants of prime interest: SLC9A4 c.1919G>A, KIAA1109 c.2933T>C and c.4268_4269delCCinsTA, HoxB6 c.668C>A, HoxD12 c.418G>A, and NCK2 c.745_746delAAinsG, from which NCK2 c.745_746delAAinsG is novel. Data validation in pediatric celiac disease patients of Greek (n = 109) and Serbian (n = 73) descent and their healthy counterparts (n = 111 and n = 32, respectively) indicated that HoxD12 c.418G>A is more prevalent in celiac disease patients in the Serbian population (P < 0.01), while NCK2 c.745_746delAAinsG is less prevalent in celiac disease patients rather than healthy individuals of Greek descent (P = 0.03). SLC9A4 c.1919G>A and KIAA1109 c.2933T>C and c.4268_4269delCCinsTA were more abundant in patients; nevertheless, they failed to show statistical significance.
Conclusions: The next-generation sequencing-based family genomics approach described herein may serve as a paradigm towards the identification of novel functional variants with the aim of understanding complex disease pathobiology.
Keywords: Celiac disease; Disease predisposition; Family genomics; Genomic variants; Next-generation sequencing.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

