DNA Topoisomerase Iα Affects the Floral Transition
- PMID: 27837087
- PMCID: PMC5210759
- DOI: 10.1104/pp.16.01603
DNA Topoisomerase Iα Affects the Floral Transition
Abstract
DNA topoisomerases modulate DNA topology to maintain chromosome superstructure and genome integrity, which is indispensable for DNA replication and RNA transcription. Their function in plant development still remains largely unknown. Here, we report a hitherto unidentified role of Topoisomerase Iα (TOP1α) in controlling flowering time in Arabidopsis (Arabidopsis thaliana). Loss of function of TOP1α results in early flowering under both long and short days. This is attributed mainly to a decrease in the expression of a central flowering repressor, FLOWERING LOCUS C (FLC), and its close homologs, MADS AFFECTING FLOWERING4 (MAF4) and MAF5, during the floral transition. TOP1α physically binds to the genomic regions of FLC, MAF4, and MAF5 and promotes the association of RNA polymerase II complexes to their transcriptional start sites. These correlate with the changes in histone modifications but do not directly affect nucleosome occupancy at these loci. Our results suggest that TOP1α mediates DNA topology to facilitate the recruitment of RNA polymerase II at FLC, MAF4, and MAF5 in conjunction with histone modifications, thus facilitating the expression of these key flowering repressors to prevent precocious flowering in Arabidopsis.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures
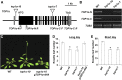
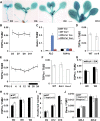

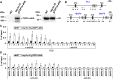
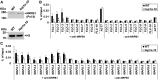

Similar articles
-
DEK domain-containing proteins control flowering time in Arabidopsis.New Phytol. 2021 Jul;231(1):182-192. doi: 10.1111/nph.17366. Epub 2021 May 2. New Phytol. 2021. PMID: 33774831 Free PMC article.
-
The putative PRC1 RING-finger protein AtRING1A regulates flowering through repressing MADS AFFECTING FLOWERING genes in Arabidopsis.Development. 2014 Mar;141(6):1303-12. doi: 10.1242/dev.104513. Epub 2014 Feb 19. Development. 2014. PMID: 24553292
-
TOP1α regulates FLOWERING LOCUS C expression by coupling histone modification and transcription machinery.Development. 2019 Feb 15;146(4):dev167841. doi: 10.1242/dev.167841. Development. 2019. PMID: 30705075
-
Just say no: floral repressors help Arabidopsis bide the time.Curr Opin Plant Biol. 2009 Oct;12(5):580-6. doi: 10.1016/j.pbi.2009.07.006. Epub 2009 Aug 18. Curr Opin Plant Biol. 2009. PMID: 19695946 Review.
-
Role of chromatin modification in flowering-time control.Trends Plant Sci. 2005 Jan;10(1):30-5. doi: 10.1016/j.tplants.2004.11.003. Trends Plant Sci. 2005. PMID: 15642521 Review.
Cited by
-
Arabidopsis DNA topoisomerase I alpha is required for adaptive response to light and flower development.Biol Open. 2017 Jun 15;6(6):832-843. doi: 10.1242/bio.024422. Biol Open. 2017. PMID: 28495963 Free PMC article.
-
DEK domain-containing proteins control flowering time in Arabidopsis.New Phytol. 2021 Jul;231(1):182-192. doi: 10.1111/nph.17366. Epub 2021 May 2. New Phytol. 2021. PMID: 33774831 Free PMC article.
-
Genome-wide dynamic network analysis reveals a critical transition state of flower development in Arabidopsis.BMC Plant Biol. 2019 Jan 7;19(1):11. doi: 10.1186/s12870-018-1589-6. BMC Plant Biol. 2019. PMID: 30616516 Free PMC article.
-
Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses.Front Plant Sci. 2018 Mar 27;9:416. doi: 10.3389/fpls.2018.00416. eCollection 2018. Front Plant Sci. 2018. PMID: 29636768 Free PMC article. Review.
-
The role of DNA topoisomerase 1α (AtTOP1α) in regulating arabidopsis meiotic recombination and chromosome segregation.PeerJ. 2024 Aug 28;12:e17864. doi: 10.7717/peerj.17864. eCollection 2024. PeerJ. 2024. PMID: 39221285 Free PMC article.
References
-
- Andersson CR, Helliwell CA, Bagnall DJ, Hughes TP, Finnegan EJ, Peacock WJ, Dennis ES (2008) The FLX gene of Arabidopsis is required for FRI-dependent activation of FLC expression. Plant Cell Physiol 49: 191–200 - PubMed
-
- Ashour ME, Atteya R, El-Khamisy SF (2015) Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat Rev Cancer 15: 137–151 - PubMed
-
- Ausín I, Alonso-Blanco C, Jarillo JA, Ruiz-García L, Martínez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36: 162–166 - PubMed
-
- Buzas DM, Robertson M, Finnegan EJ, Helliwell CA (2011) Transcription-dependence of histone H3 lysine 27 trimethylation at the Arabidopsis polycomb target gene FLC. Plant J 65: 872–881 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

