Membrane Proteomic Insights into the Physiology and Taxonomy of an Oleaginous Green Microalga
- PMID: 27837088
- PMCID: PMC5210721
- DOI: 10.1104/pp.16.01240
Membrane Proteomic Insights into the Physiology and Taxonomy of an Oleaginous Green Microalga
Abstract
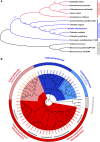
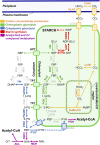
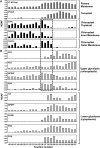
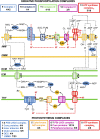
Ettlia oleoabundans is a nonsequenced oleaginous green microalga. Despite the significant biotechnological interest in producing value-added compounds from the acyl lipids of this microalga, a basic understanding of the physiology and biochemistry of oleaginous microalgae is lacking, especially under nitrogen deprivation conditions known to trigger lipid accumulation. Using an RNA sequencing-based proteomics approach together with manual annotation, we are able to provide, to our knowledge, the first membrane proteome of an oleaginous microalga. This approach allowed the identification of novel proteins in E. oleoabundans, including two photoprotection-related proteins, Photosystem II Subunit S and Maintenance of Photosystem II under High Light1, which were considered exclusive to higher photosynthetic organisms, as well as Retinitis Pigmentosa Type 2-Clathrin Light Chain, a membrane protein with a novel domain architecture. Free-flow zonal electrophoresis of microalgal membranes coupled to liquid chromatography-tandem mass spectrometry proved to be a useful technique for determining the intracellular location of proteins of interest. Carbon-flow compartmentalization in E. oleoabundans was modeled using this information. Molecular phylogenetic analyses of protein markers and 18S ribosomal DNA support the reclassification of E. oleoabundans within the trebouxiophycean microalgae, rather than with the Chlorophyceae class, in which it is currently classified, indicating that it may not be closely related to the model green alga Chlamydomonas reinhardtii A detailed survey of biological processes taking place in the membranes of nitrogen-deprived E. oleoabundans, including lipid metabolism, provides insights into the basic biology of this nonmodel organism.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures
Similar articles
-
Time-series lipidomic analysis of the oleaginous green microalga species Ettlia oleoabundans under nutrient stress.Biotechnol Biofuels. 2018 Feb 6;11:29. doi: 10.1186/s13068-018-1026-y. eCollection 2018. Biotechnol Biofuels. 2018. PMID: 29441127 Free PMC article.
-
Comparative proteomics reveals proteins impacted by nitrogen deprivation in wild-type and high lipid-accumulating mutant strains of Tisochrysis lutea.J Proteomics. 2014 Jun 13;105:107-20. doi: 10.1016/j.jprot.2014.02.022. Epub 2014 Feb 28. J Proteomics. 2014. PMID: 24583506
-
Comparative Shotgun Proteomic Analysis of Wastewater-Cultured Microalgae: Nitrogen Sensing and Carbon Fixation for Growth and Nutrient Removal in Chlamydomonas reinhardtii.J Proteome Res. 2015 Aug 7;14(8):3051-67. doi: 10.1021/pr501316h. Epub 2015 Jul 8. J Proteome Res. 2015. PMID: 25997359
-
Carbon acquisition and accumulation in microalgae Chlamydomonas: Insights from "omics" approaches.J Proteomics. 2013 Dec 6;94:207-18. doi: 10.1016/j.jprot.2013.09.016. Epub 2013 Oct 11. J Proteomics. 2013. PMID: 24120529 Review.
-
The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features.Curr Genet. 2011 Jun;57(3):151-68. doi: 10.1007/s00294-011-0339-1. Epub 2011 Apr 30. Curr Genet. 2011. PMID: 21533645 Review.
Cited by
-
Marine Natural Products from Microalgae: An -Omics Overview.Mar Drugs. 2019 May 7;17(5):269. doi: 10.3390/md17050269. Mar Drugs. 2019. PMID: 31067655 Free PMC article. Review.
-
Comparative Proteomics Reveals Evidence of Enhanced EPA Trafficking in a Mutant Strain of Nannochloropsis oculata.Front Bioeng Biotechnol. 2022 May 12;10:838445. doi: 10.3389/fbioe.2022.838445. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35646838 Free PMC article.
-
Shotgun proteomics of Brassica rapa seed proteins identifies vicilin as a major seed storage protein in the mature seed.PLoS One. 2021 Jul 9;16(7):e0253384. doi: 10.1371/journal.pone.0253384. eCollection 2021. PLoS One. 2021. PMID: 34242257 Free PMC article.
-
Development of New Antiproliferative Compound against Human Tumor Cells from the Marine Microalgae Nannochloropsis gaditana by Applied Proteomics.Int J Mol Sci. 2020 Dec 24;22(1):96. doi: 10.3390/ijms22010096. Int J Mol Sci. 2020. PMID: 33374179 Free PMC article.
-
Light and Dehydration but Not Temperature Drive Photosynthetic Adaptations of Basal Streptophytes (Hormidiella, Streptosarcina and Streptofilum) Living in Terrestrial Habitats.Microb Ecol. 2019 Feb;77(2):380-393. doi: 10.1007/s00248-018-1225-x. Epub 2018 Jul 4. Microb Ecol. 2019. PMID: 29974184 Free PMC article.
References
-
- Arredondo-Vega BO, Band CJ, Vazquez-Duhalt R (1995) Biochemical composition of Neochloris oleoabundans adapted to marine medium. Cytobios 83: 201–205
-
- Atteia A, Adrait A, Brugière S, Tardif M, van Lis R, Deusch O, Dagan T, Kuhn L, Gontero B, Martin W, et al. (2009) A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-proteobacterial mitochondrial ancestor. Mol Biol Evol 26: 1533–1548 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

