Transcriptome profiling of NIH3T3 cell lines expressing opsin and the P23H opsin mutant identifies candidate drugs for the treatment of retinitis pigmentosa
- PMID: 27838510
- PMCID: PMC5205556
- DOI: 10.1016/j.phrs.2016.10.031
Transcriptome profiling of NIH3T3 cell lines expressing opsin and the P23H opsin mutant identifies candidate drugs for the treatment of retinitis pigmentosa
Abstract
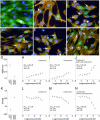
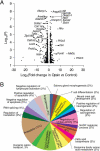
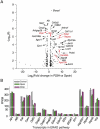
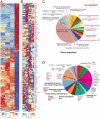
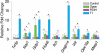
Mammalian cells are commonly employed in screening assays to identify active compounds that could potentially affect the progression of different human diseases including retinitis pigmentosa (RP), a class of inherited diseases causing retinal degeneration with compromised vision. Using transcriptome analysis, we compared NIH3T3 cells expressing wildtype (WT) rod opsin with a retinal disease-causing single P23H mutation. Surprisingly, heterologous expression of WT opsin in NIH3T3 cells caused more than a 2-fold change in 783 out of 16,888 protein coding transcripts. The perturbed genes encoded extracellular matrix proteins, growth factors, cytoskeleton proteins, glycoproteins and metalloproteases involved in cell adhesion, morphology and migration. A different set of 347 transcripts was either up- or down-regulated when the P23H mutant opsin was expressed suggesting an altered molecular perturbation compared to WT opsin. Transcriptome perturbations elicited by drug candidates aimed at mitigating the effects of the mutant protein revealed that different drugs targeted distinct molecular pathways that resulted in a similar phenotype selected by a cell-based high-throughput screen. Thus, transcriptome profiling can provide essential information about the therapeutic potential of a candidate drug to restore normal gene expression in pathological conditions.
Keywords: Cell-based HTS; Drug discovery; P23H opsin; RNA-seq; Retina; Retinitis pigmentosa; Rhodopsin; Transcriptome.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
P23H opsin knock-in mice reveal a novel step in retinal rod disc morphogenesis.Hum Mol Genet. 2014 Apr 1;23(7):1723-41. doi: 10.1093/hmg/ddt561. Epub 2013 Nov 7. Hum Mol Genet. 2014. PMID: 24214395 Free PMC article.
-
Inherent instability of the retinitis pigmentosa P23H mutant opsin.J Biol Chem. 2014 Mar 28;289(13):9288-303. doi: 10.1074/jbc.M114.551713. Epub 2014 Feb 10. J Biol Chem. 2014. PMID: 24515108 Free PMC article.
-
Retinoids assist the cellular folding of the autosomal dominant retinitis pigmentosa opsin mutant P23H.J Biol Chem. 2004 Apr 16;279(16):16278-84. doi: 10.1074/jbc.M312101200. Epub 2004 Feb 9. J Biol Chem. 2004. PMID: 14769795
-
Pharmacological manipulation of rhodopsin retinitis pigmentosa.Adv Exp Med Biol. 2010;664:317-23. doi: 10.1007/978-1-4419-1399-9_36. Adv Exp Med Biol. 2010. PMID: 20238031 Review.
-
Targeting the Proteostasis Network in Rhodopsin Retinitis Pigmentosa.Adv Exp Med Biol. 2016;854:479-84. doi: 10.1007/978-3-319-17121-0_64. Adv Exp Med Biol. 2016. PMID: 26427449 Free PMC article. Review.
Cited by
-
A novel small molecule chaperone of rod opsin and its potential therapy for retinal degeneration.Nat Commun. 2018 May 17;9(1):1976. doi: 10.1038/s41467-018-04261-1. Nat Commun. 2018. PMID: 29773803 Free PMC article.
-
Functional Genomics of the Retina to Elucidate its Construction and Deconstruction.Int J Mol Sci. 2019 Oct 4;20(19):4922. doi: 10.3390/ijms20194922. Int J Mol Sci. 2019. PMID: 31590277 Free PMC article. Review.
-
RNA-sequencing in ophthalmology research: considerations for experimental design and analysis.Ther Adv Ophthalmol. 2019 Mar 15;11:2515841419835460. doi: 10.1177/2515841419835460. eCollection 2019 Jan-Dec. Ther Adv Ophthalmol. 2019. PMID: 30911735 Free PMC article. Review.
-
A Rhodopsin Transport Assay by High-Content Imaging Analysis.J Vis Exp. 2019 Jan 16;(143):10.3791/58703. doi: 10.3791/58703. J Vis Exp. 2019. PMID: 30735172 Free PMC article.
References
-
- Westerhoff HV, Nakayama S, Mondeel TD, Barberis M. Systems Pharmacology: An opinion on how to turn the impossible into grand challenges. Drug discovery today. Technologies. 2015;15:23–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

