Complex Ancient Genetic Structure and Cultural Transitions in Southern African Populations
- PMID: 27838627
- PMCID: PMC5223510
- DOI: 10.1534/genetics.116.189209
Complex Ancient Genetic Structure and Cultural Transitions in Southern African Populations
Abstract
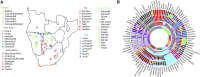
The characterization of the structure of southern African populations has been the subject of numerous genetic, medical, linguistic, archaeological, and anthropological investigations. Current diversity in the subcontinent is the result of complex events of genetic admixture and cultural contact between early inhabitants and migrants that arrived in the region over the last 2000 years. Here, we analyze 1856 individuals from 91 populations, comprising novel and published genotype data, to characterize the genetic ancestry profiles of 631 individuals from 51 southern African populations. Combining both local ancestry and allele frequency based analyses, we identify a tripartite, ancient, Khoesan-related genetic structure. This structure correlates neither with linguistic affiliation nor subsistence strategy, but with geography, revealing the importance of isolation-by-distance dynamics in the area. Fine-mapping of these components in southern African populations reveals admixture and cultural reversion involving several Khoesan groups, and highlights that Bantu speakers and Coloured individuals have different mixtures of these ancient ancestries.
Keywords: African prehistory; Khoesan; ancient structure; sub-Saharan Africa.
Copyright © 2017 Montinaro et al.
Figures


References
-
- Barham L., Mitchell P., 2008. The First Africans: African Archaeology from the Earliest Toolmakers to Most Recent Foragers. Cambridge University Press, Cambridge, UK.
-
- Barnard A., 1992. Hunters and Herders of Southern Africa: A Comparative Ethnography of the Khoisan Peoples. Cambridge University Press, Cambridge, UK.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

