Haploinsufficiency of KMT2B, Encoding the Lysine-Specific Histone Methyltransferase 2B, Results in Early-Onset Generalized Dystonia
- PMID: 27839873
- PMCID: PMC5142117
- DOI: 10.1016/j.ajhg.2016.10.010
Haploinsufficiency of KMT2B, Encoding the Lysine-Specific Histone Methyltransferase 2B, Results in Early-Onset Generalized Dystonia
Abstract
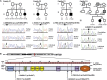
Early-onset generalized dystonia represents the severest form of dystonia, a hyperkinetic movement disorder defined by involuntary twisting postures. Although frequently transmitted as a single-gene trait, the molecular basis of dystonia remains largely obscure. By whole-exome sequencing a parent-offspring trio in an Austrian kindred affected by non-familial early-onset generalized dystonia, we identified a dominant de novo frameshift mutation, c.6406delC (p.Leu2136Serfs∗17), in KMT2B, encoding a lysine-specific methyltransferase involved in transcriptional regulation via post-translational modification of histones. Whole-exome-sequencing-based exploration of a further 30 German-Austrian individuals with early-onset generalized dystonia uncovered another three deleterious mutations in KMT2B-one de novo nonsense mutation (c.1633C>T [p.Arg545∗]), one de novo essential splice-site mutation (c.7050-2A>G [p.Phe2321Serfs∗93]), and one inherited nonsense mutation (c.2428C>T [p.Gln810∗]) co-segregating with dystonia in a three-generation kindred. Each of the four mutations was predicted to mediate a loss-of-function effect by introducing a premature termination codon. Suggestive of haploinsufficiency, we found significantly decreased total mRNA levels of KMT2B in mutant fibroblasts. The phenotype of individuals with KMT2B loss-of-function mutations was dominated by childhood lower-limb-onset generalized dystonia, and the family harboring c.2428C>T (p.Gln810∗) showed variable expressivity. In most cases, dystonic symptoms were accompanied by heterogeneous non-motor features. Independent support for pathogenicity of the mutations comes from the observation of high rates of dystonic presentations in KMT2B-involving microdeletion syndromes. Our findings thus establish generalized dystonia as the human phenotype associated with haploinsufficiency of KMT2B. Moreover, we provide evidence for a causative role of disordered histone modification, chromatin states, and transcriptional deregulation in dystonia pathogenesis.
Copyright © 2016 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


Comment in
-
New gene implicated in early-onset generalized dystonia: Lysine-specific methyltransferase 2B (KMT2B).Mov Disord. 2017 Mar;32(3):395. doi: 10.1002/mds.26928. Epub 2017 Feb 2. Mov Disord. 2017. PMID: 28150430 No abstract available.
-
KMT2B: A new twist in dystonia genetics.Mov Disord. 2017 Apr;32(4):529. doi: 10.1002/mds.26957. Epub 2017 Feb 20. Mov Disord. 2017. PMID: 28218417 No abstract available.
References
-
- Balint B., Bhatia K.P. Dystonia: an update on phenomenology, classification, pathogenesis and treatment. Curr. Opin. Neurol. 2014;27:468–476. - PubMed
-
- Klein C. Genetics in dystonia. Parkinsonism Relat. Disord. 2014;20(Suppl 1):S137–S142. - PubMed
-
- Domingo A., Erro R., Lohmann K. Novel Dystonia Genes: Clues on Disease Mechanisms and the Complexities of High-Throughput Sequencing. Mov. Disord. 2016;31:471–477. - PubMed
-
- Zech M., Boesch S., Jochim A., Weber S., Meindl T., Schormair B., Wieland T., Lunetta C., Sansone V., Messner M. Clinical exome sequencing in early-onset generalized dystonia and large-scale resequencing follow-up. Mov. Disord. 2016 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

