Live Monitoring of Blastemal Cell Contributions during Appendage Regeneration
- PMID: 27839971
- PMCID: PMC5121098
- DOI: 10.1016/j.cub.2016.08.072
Live Monitoring of Blastemal Cell Contributions during Appendage Regeneration
Abstract
The blastema is a mass of progenitor cells that enables regeneration of amputated salamander limbs or fish fins. Methodology to label and track blastemal cell progeny has been deficient, restricting our understanding of appendage regeneration. Here, we created a system for clonal analysis and quantitative imaging of hundreds of blastemal cells and their respective progeny in living adult zebrafish undergoing fin regeneration. Amputation stimulates resident cells within a limited recruitment zone to reset proximodistal (PD) positional information and assemble the blastema. Within the newly formed blastema, the spatial coordinates of connective tissue progenitors are predictive of their ultimate contributions to regenerated skeletal structures, indicating early development of an approximate PD pre-pattern. Calcineurin regulates size recovery by controlling the average number of progeny divisions without disrupting this pre-pattern. Our longitudinal clonal analyses of regenerating zebrafish fins provide evidence that connective tissue progenitors are rapidly organized into a scalable blueprint of lost structures.
Keywords: appendage; blastema; clonal analysis; fibroblast; fin; positional identity; regeneration; tph1b; vertebrate; zebrafish.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
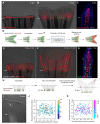
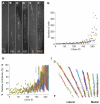
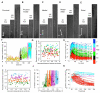
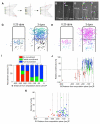


Comment in
-
Regeneration: Recorded Live!Curr Biol. 2017 Jan 9;27(1):R30-R33. doi: 10.1016/j.cub.2016.11.022. Curr Biol. 2017. PMID: 28073020
References
-
- Simon A, Tanaka EM. Limb regeneration. Wiley Interdiscip Rev Dev Biol. 2013;2:291–300. - PubMed
-
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. - PubMed
-
- Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S, et al. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Developmental Cell. 2011;20:713–724. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

