Archaic Hominin Admixture Facilitated Adaptation to Out-of-Africa Environments
- PMID: 27839976
- PMCID: PMC6764441
- DOI: 10.1016/j.cub.2016.10.041
Archaic Hominin Admixture Facilitated Adaptation to Out-of-Africa Environments
Abstract
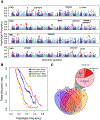
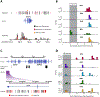
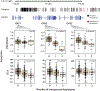
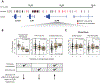
As modern humans dispersed from Africa throughout the world, they encountered and interbred with archaic hominins, including Neanderthals and Denisovans [1, 2]. Although genome-scale maps of introgressed sequences have been constructed [3-6], considerable gaps in knowledge remain about the functional, phenotypic, and evolutionary significance of archaic hominin DNA that persists in present-day individuals. Here, we describe a comprehensive set of analyses that identified 126 high-frequency archaic haplotypes as putative targets of adaptive introgression in geographically diverse populations. These loci are enriched for immune-related genes (such as OAS1/2/3, TLR1/6/10, and TNFAIP3) and also encompass genes (including OCA2 and BNC2) that influence skin pigmentation phenotypes. Furthermore, we leveraged existing and novel large-scale gene expression datasets to show many positively selected archaic haplotypes act as expression quantitative trait loci (eQTLs), suggesting that modulation of transcript abundance was a common mechanism facilitating adaptive introgression. Our results demonstrate that hybridization between modern and archaic hominins provided an important reservoir of advantageous alleles that enabled adaptation to out-of-Africa environments.
Keywords: Denisovan; Hybridization; Neandertal; adaptive; admixture; human evolution; introgression.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Something old, something borrowed: admixture and adaptation in human evolution.Curr Opin Genet Dev. 2018 Dec;53:1-8. doi: 10.1016/j.gde.2018.05.009. Epub 2018 Jun 9. Curr Opin Genet Dev. 2018. PMID: 29894925 Review.
-
Archaic Introgression Shaped Human Circadian Traits.Genome Biol Evol. 2023 Dec 1;15(12):evad203. doi: 10.1093/gbe/evad203. Genome Biol Evol. 2023. PMID: 38095367 Free PMC article.
-
Archaic hominin admixture and its consequences for modern humans.Curr Opin Genet Dev. 2025 Feb;90:102280. doi: 10.1016/j.gde.2024.102280. Epub 2024 Nov 21. Curr Opin Genet Dev. 2025. PMID: 39577372 Review.
-
Introgression of Neandertal- and Denisovan-like Haplotypes Contributes to Adaptive Variation in Human Toll-like Receptors.Am J Hum Genet. 2016 Jan 7;98(1):22-33. doi: 10.1016/j.ajhg.2015.11.015. Am J Hum Genet. 2016. PMID: 26748514 Free PMC article.
-
Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals.Science. 2016 Apr 8;352(6282):235-9. doi: 10.1126/science.aad9416. Epub 2016 Mar 17. Science. 2016. PMID: 26989198 Free PMC article.
Cited by
-
Neanderthal introgression reintroduced functional ancestral alleles lost in Eurasian populations.Nat Ecol Evol. 2020 Oct;4(10):1332-1341. doi: 10.1038/s41559-020-1261-z. Epub 2020 Jul 27. Nat Ecol Evol. 2020. PMID: 32719451 Free PMC article.
-
An approximate full-likelihood method for inferring selection and allele frequency trajectories from DNA sequence data.PLoS Genet. 2019 Sep 13;15(9):e1008384. doi: 10.1371/journal.pgen.1008384. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31518343 Free PMC article.
-
The genomic signatures of natural selection in admixed human populations.Am J Hum Genet. 2022 Apr 7;109(4):710-726. doi: 10.1016/j.ajhg.2022.02.011. Epub 2022 Mar 7. Am J Hum Genet. 2022. PMID: 35259336 Free PMC article.
-
The Impact of Recessive Deleterious Variation on Signals of Adaptive Introgression in Human Populations.Genetics. 2020 Jul;215(3):799-812. doi: 10.1534/genetics.120.303081. Epub 2020 Jun 2. Genetics. 2020. PMID: 32487519 Free PMC article.
-
The role of genetic selection and climatic factors in the dispersal of anatomically modern humans out of Africa.Proc Natl Acad Sci U S A. 2023 May 30;120(22):e2213061120. doi: 10.1073/pnas.2213061120. Epub 2023 May 23. Proc Natl Acad Sci U S A. 2023. PMID: 37220274 Free PMC article.
References
-
- Vernot B, Akey JM (2014) Resurrecting surviving Neandertal lineages from modern human genomes. Science. 343, 1017–1021. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

