CRY2 and FBXL3 Cooperatively Degrade c-MYC
- PMID: 27840026
- PMCID: PMC5123859
- DOI: 10.1016/j.molcel.2016.10.012
CRY2 and FBXL3 Cooperatively Degrade c-MYC
Abstract
For many years, a connection between circadian clocks and cancer has been postulated. Here we describe an unexpected function for the circadian repressor CRY2 as a component of an FBXL3-containing E3 ligase that recruits T58-phosphorylated c-MYC for ubiquitylation. c-MYC is a critical regulator of cell proliferation; T58 is central in a phosphodegron long recognized as a hotspot for mutation in cancer. This site is also targeted by FBXW7, although the full machinery responsible for its turnover has remained obscure. CRY1 cannot substitute for CRY2 in promoting c-MYC degradation. Their unique functions may explain prior conflicting reports that have fueled uncertainty about the relationship between clocks and cancer. We demonstrate that c-MYC is a target of CRY2-dependent protein turnover, suggesting a molecular mechanism for circadian control of cell growth and a new paradigm for circadian protein degradation.
Keywords: CRY2; FBXL3; MYC; circadian clock; circadian rhythm; cryptochrome; ubiquitin.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
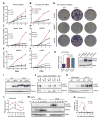
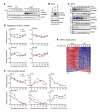

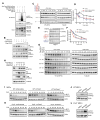

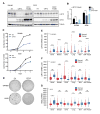
References
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. - PubMed
-
- Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt’s lymphoma and mouse plasmacytomas. Nature genetics. 1993;5:56–61. - PubMed
-
- Bhatia K, Spangler G, Gaidano G, Hamdy N, Dalla-Favera R, Magrath I. Mutations in the coding region of c-myc occur frequently in acquired immunodeficiency syndrome-associated lymphomas. Blood. 1994;84:883–888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

