HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling
- PMID: 27841315
- PMCID: PMC5107982
- DOI: 10.1038/srep37021
HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling
Abstract
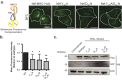
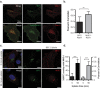
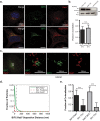
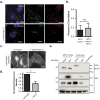
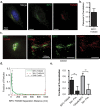
A defining characteristic of HIV-1 infection is the ability of the virus to persist within the host. Specifically, MHC-I downregulation by the HIV-1 accessory protein Nef is of critical importance in preventing infected cells from cytotoxic T-cell mediated killing. Nef downregulates MHC-I by modulating the host membrane trafficking machinery, resulting in the endocytosis and eventual sequestration of MHC-I within the cell. In the current report, we utilized the intracellular protein-protein interaction reporter system, bimolecular fluorescence complementation (BiFC), in combination with super-resolution microscopy, to track the Nef/MHC-I interaction and determine its subcellular localization in cells. We demonstrate that this interaction occurs upon Nef binding the MHC-I cytoplasmic tail early during endocytosis in a Rab5-positive endosome. Disruption of early endosome regulation inhibited Nef-dependent MHC-I downregulation, demonstrating that Nef hijacks the early endosome to sequester MHC-I within the cell. Furthermore, super-resolution imaging identified that the Nef:MHC-I BiFC complex transits through both early and late endosomes before ultimately residing at the trans-Golgi network. Together we demonstrate the importance of the early stages of the endocytic network in the removal of MHC-I from the cell surface and its re-localization within the cell, which allows HIV-1 to optimally evade host immune responses.
Figures

References
-
- Malim M. H. & Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398, doi: S1931–3128(08)00126-1 (2008). - PubMed
-
- Blagoveshchenskaya A. D., Thomas L., Feliciangeli S. F., Hung C. H. & Thomas G. HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111, 853–866, doi: S0092867402011625 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

