Genetic architecture of the maize kernel row number revealed by combining QTL mapping using a high-density genetic map and bulked segregant RNA sequencing
- PMID: 27842488
- PMCID: PMC5109822
- DOI: 10.1186/s12864-016-3240-y
Genetic architecture of the maize kernel row number revealed by combining QTL mapping using a high-density genetic map and bulked segregant RNA sequencing
Abstract
Background: The maize kernel row number (KRN) is a key component that contributes to grain yield and has high broad-sense heritability (H 2 ). Quantitative trait locus/loci (QTL) mapping using a high-density genetic map is a powerful approach to detecting loci that are responsible for traits of interest. Bulked segregant ribonucleic acid (RNA) sequencing (BSR-seq) is another rapid and cost-effective strategy to identify QTL. Combining QTL mapping using a high-density genetic map and BSR-seq may dissect comprehensively the genetic architecture underlying the maize KRN.
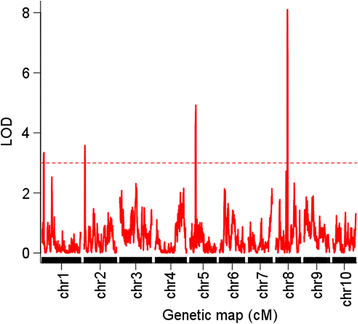
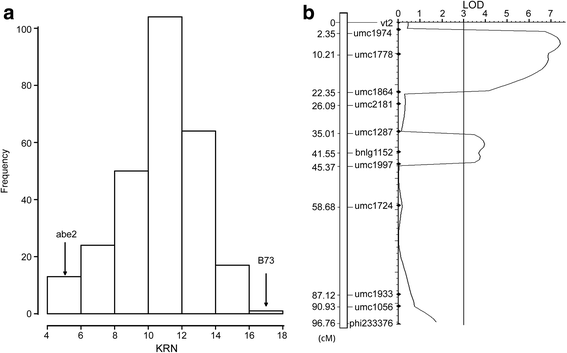
Results: A panel of 300 F2 individuals derived from inbred lines abe2 and B73 were genotyped using the specific-locus amplified fragment sequencing (SLAF-seq) method. A total of 4,579 high-quality polymorphic SLAF markers were obtained and used to construct a high-density genetic map with a total length of 2,123 centimorgan (cM) and an average distance between adjacent markers of 0.46 cM. Combining the genetic map and KRN of F2 individuals, four QTL (qKRN1, qKRN2, qKRN5, and qKRN8-1) were identified on chromosomes 1, 2, 5, and 8, respectively. The physical intervals of these four QTL ranged from 4.36 Mb for qKRN8-1 to 7.11 Mb for qKRN1 with an average value of 6.08 Mb. Based on high-throughput sequencing of two RNA pools bulked from leaves of plants with extremely high and low KRNs, two QTL were detected on chromosome 8 in the 10-25 Mb (BSR_QTL1) and 60-150 Mb (BSR_QTL2) intervals. According to the physical positions of these QTL, qKRN8-1 was included by BSR_QTL2. In addition, qKRN8-1 was validated using QTL mapping with a recombinant inbred lines population that was derived from inbred lines abe2 and B73.
Conclusions: In this study, we proved that combining QTL mapping using a high-density genetic map and BSR-seq is a powerful and cost-effective approach to comprehensively revealing genetic architecture underlying traits of interest. The QTL for the KRN detected in this study, especially qKRN8-1, can be used for performing fine mapping experiments and marker-assisted selection in maize breeding.
Keywords: Bulked segregant RNA sequencing; Kernel row number; Maize; Specific-locus amplified fragment sequencing.
Figures


Similar articles
-
Genetic dissection of maize plant architecture with an ultra-high density bin map based on recombinant inbred lines.BMC Genomics. 2016 Mar 3;17:178. doi: 10.1186/s12864-016-2555-z. BMC Genomics. 2016. PMID: 26940065 Free PMC article.
-
Fine mapping of qKRN8, a QTL for maize kernel row number, and prediction of the candidate gene.Theor Appl Genet. 2020 Nov;133(11):3139-3150. doi: 10.1007/s00122-020-03660-7. Epub 2020 Aug 28. Theor Appl Genet. 2020. PMID: 32857170
-
Construction of a high-density genetic map based on large-scale markers developed by specific length amplified fragment sequencing (SLAF-seq) and its application to QTL analysis for isoflavone content in Glycine max.BMC Genomics. 2014 Dec 10;15(1):1086. doi: 10.1186/1471-2164-15-1086. BMC Genomics. 2014. PMID: 25494922 Free PMC article.
-
Review: Recent advances in unraveling the genetic architecture of kernel row number in maize.Plant Sci. 2025 Mar;352:112366. doi: 10.1016/j.plantsci.2024.112366. Epub 2024 Dec 20. Plant Sci. 2025. PMID: 39710150 Review.
-
Hotspot Regions of Quantitative Trait Loci and Candidate Genes for Ear-Related Traits in Maize: A Literature Review.Genes (Basel). 2023 Dec 21;15(1):15. doi: 10.3390/genes15010015. Genes (Basel). 2023. PMID: 38275597 Free PMC article. Review.
Cited by
-
Identification of fruit size associated quantitative trait loci featuring SLAF based high-density linkage map of goji berry (Lycium spp.).BMC Plant Biol. 2020 Oct 15;20(1):474. doi: 10.1186/s12870-020-02567-1. BMC Plant Biol. 2020. PMID: 33059596 Free PMC article.
-
Predicting Zea mays Flowering Time, Yield, and Kernel Dimensions by Analyzing Aerial Images.Front Plant Sci. 2019 Oct 11;10:1251. doi: 10.3389/fpls.2019.01251. eCollection 2019. Front Plant Sci. 2019. PMID: 31681364 Free PMC article.
-
Genetic Architecture of Grain Yield-Related Traits in Sorghum and Maize.Int J Mol Sci. 2022 Feb 22;23(5):2405. doi: 10.3390/ijms23052405. Int J Mol Sci. 2022. PMID: 35269548 Free PMC article. Review.
-
Phosphorus partitioning contribute to phosphorus use efficiency during grain filling in Zea mays.Front Plant Sci. 2023 Jul 4;14:1223532. doi: 10.3389/fpls.2023.1223532. eCollection 2023. Front Plant Sci. 2023. PMID: 37469778 Free PMC article.
-
Identification and Validation of New Stable QTLs for Grain Weight and Size by Multiple Mapping Models in Common Wheat.Front Genet. 2020 Nov 11;11:584859. doi: 10.3389/fgene.2020.584859. eCollection 2020. Front Genet. 2020. PMID: 33262789 Free PMC article.
References
-
- Qin X, Feng F, Li Y, Xu S, Siddique KHM, Liao Y, Lübberstedt T. Maize yield improvements in China: past trends and future directions. Plant Breed. 2016;135:166–76. doi: 10.1111/pbr.12347. - DOI
-
- Yang C, Tang D, Zhang L, Liu J, Rong T. Identification of QTL for ear row number and two-ranked versus many-ranked ear in maize across four environments. Euphytica. 2015;206:33–47. doi: 10.1007/s10681-015-1466-4. - DOI
-
- Lu M, Xie CX, Li XH, Hao ZF, Li MS, Weng JF, Zhang DG, Bai L, Zhang SH. Mapping of quantitative trait loci for kernel row number in maize across seven environments. Mol Breeding. 2010;28:143–52. doi: 10.1007/s11032-010-9468-3. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

