A Decade-Long Commitment to Antimicrobial Resistance Surveillance in Portugal
- PMID: 27843438
- PMCID: PMC5086874
- DOI: 10.3389/fmicb.2016.01650
A Decade-Long Commitment to Antimicrobial Resistance Surveillance in Portugal
Abstract
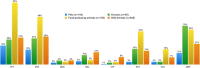
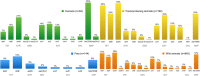
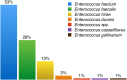
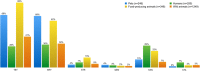
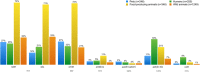
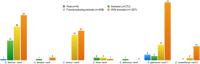
Antimicrobial resistance (AMR) is a worldwide problem with serious health and economic repercussions. Since the 1940s, underuse, overuse, and misuse of antibiotics have had a significant environmental downside. Large amounts of antibiotics not fully metabolized after use in human and veterinary medicine, and other applications, are annually released into the environment. The result has been the development and dissemination of antibiotic-resistant bacteria due to many years of selective pressure. Surveillance of AMR provides important information that helps in monitoring and understanding how resistance mechanisms develop and disseminate within different environments. Surveillance data is needed to inform clinical therapy decisions, to guide policy proposals, and to assess the impact of action plans to fight AMR. The Functional Genomics and Proteomics Unit, based at the University of Trás-os-Montes and Alto Douro in Vila Real, Portugal, has recently completed 10 years of research surveying AMR in bacteria, mainly commensal indicator bacteria such as enterococci and Escherichia coli from the microbiota of different animals. Samples from more than 75 different sources have been accessed, from humans to food-producing animals, pets, and wild animals. The typical microbiological workflow involved phenotypic studies followed by molecular approaches. Throughout the decade, 4,017 samples were collected and over 5,000 bacterial isolates obtained. High levels of AMR to several antimicrobial classes have been reported, including to β-lactams, glycopeptides, tetracyclines, aminoglycosides, sulphonamides, and quinolones. Multi-resistant strains, some relevant to human and veterinary medicine like extended-spectrum β-lactamase-producing E. coli and vancomycin-resistant enterococci, have been repeatedly isolated even in non-synanthropic animal species. Of particular relevance are reports of AMR bacteria in wildlife from natural reserves and endangered species. Future work awaits as this threatening yet unsolved problem persists. GRAPHICAL ABSTRACTSummary diagram of the antimicrobial resistance surveillance work developed by the UTAD Functional Genomics and Proteomics Unit.
Keywords: Escherichia coli; antimicrobial resistance; enterococci; molecular microbiology; surveillance; wildlife.
Figures


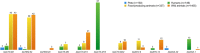
Similar articles
-
Wildlife as Sentinels of Antimicrobial Resistance in Germany?Front Vet Sci. 2021 Jan 27;7:627821. doi: 10.3389/fvets.2020.627821. eCollection 2020. Front Vet Sci. 2021. PMID: 33585611 Free PMC article.
-
Antimicrobial use and antimicrobial resistance trends in Canada: 2014.Can Commun Dis Rep. 2016 Nov 3;42(11):227-231. doi: 10.14745/ccdr.v42i11a02. eCollection 2016 Nov 3. Can Commun Dis Rep. 2016. PMID: 29769991 Free PMC article.
-
Antimicrobial resistance in Shiga toxin-producing Escherichia coli other than serotype O157 : H7 in England, 2014-2016.J Med Microbiol. 2020 Mar;69(3):379-386. doi: 10.1099/jmm.0.001146. J Med Microbiol. 2020. PMID: 32101158
-
Review of antimicrobial resistance surveillance programmes in livestock and meat in EU with focus on humans.Clin Microbiol Infect. 2018 Jun;24(6):577-590. doi: 10.1016/j.cmi.2017.09.013. Epub 2017 Sep 29. Clin Microbiol Infect. 2018. PMID: 28970159 Review.
-
Genetic diversity and risk factors for the transmission of antimicrobial resistance across human, animals and environmental compartments in East Africa: a review.Antimicrob Resist Infect Control. 2020 Aug 6;9(1):127. doi: 10.1186/s13756-020-00786-7. Antimicrob Resist Infect Control. 2020. PMID: 32762743 Free PMC article. Review.
Cited by
-
Multidrug-Resistant Phenotypes of Escherichia coli Isolates in Wild Canarian Egyptian Vultures (Neophron percnopterus majorensis).Animals (Basel). 2021 Jun 6;11(6):1692. doi: 10.3390/ani11061692. Animals (Basel). 2021. PMID: 34204084 Free PMC article.
-
Occurrence and Antimicrobial Resistance Traits of Escherichia coli from Wild Birds and Rodents in Singapore.Int J Environ Res Public Health. 2020 Aug 3;17(15):5606. doi: 10.3390/ijerph17155606. Int J Environ Res Public Health. 2020. PMID: 32756497 Free PMC article.
-
Antimicrobial Susceptibility of Enterococcus Isolates from Cattle and Pigs in Portugal: Linezolid Resistance Genes optrA and poxtA.Antibiotics (Basel). 2022 May 3;11(5):615. doi: 10.3390/antibiotics11050615. Antibiotics (Basel). 2022. PMID: 35625259 Free PMC article.
-
The Antibiotics Used in Livestock and Their Impact on Resistance in Enterococcus faecium and Enterococcus hirae on Farms in Gabon.Antibiotics (Basel). 2022 Feb 10;11(2):224. doi: 10.3390/antibiotics11020224. Antibiotics (Basel). 2022. PMID: 35203826 Free PMC article.
-
Antimicrobial resistance profiles of Escherichia coli isolated from laying hens in Zambia: implications and significance on one health.JAC Antimicrob Resist. 2023 May 22;5(3):dlad060. doi: 10.1093/jacamr/dlad060. eCollection 2023 Jun. JAC Antimicrob Resist. 2023. PMID: 37223392 Free PMC article.
References
-
- Alali W. Q., Scott H. M., Christian K. L., Fajt V. R., Harvey R. B., Lawhorn D. B. (2009). Relationship between level of antibiotic use and resistance among Escherichia coli isolates from integrated multi-site cohorts of humans and swine. Prev. Vet. Med. 90 160–167. 10.1016/j.prevetmed.2009.05.018 - DOI - PubMed
-
- Allen S. E., Janecko N., Pearl D. L., Boerlin P., Reid-Smith R. J., Jardine C. M. (2013). Comparison of Escherichia coli recovery and antimicrobial resistance in cecal, colon, and fecal samples collected from wild house mice (Mus musculus). J. Wildl. Dis. 49 432–436. 10.7589/2012-05-142 - DOI - PubMed
-
- Almeida A., Duarte S., Nunes R., Rocha H., Pena A., Meisel L. (2014). Human and veterinary antibiotics used in Portugal—a ranking for ecosurveillance. Toxics 2 188–225. 10.3390/toxics2020188 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

