Somatic retrotransposition is infrequent in glioblastomas
- PMID: 27843500
- PMCID: PMC5105304
- DOI: 10.1186/s13100-016-0077-5
Somatic retrotransposition is infrequent in glioblastomas
Abstract
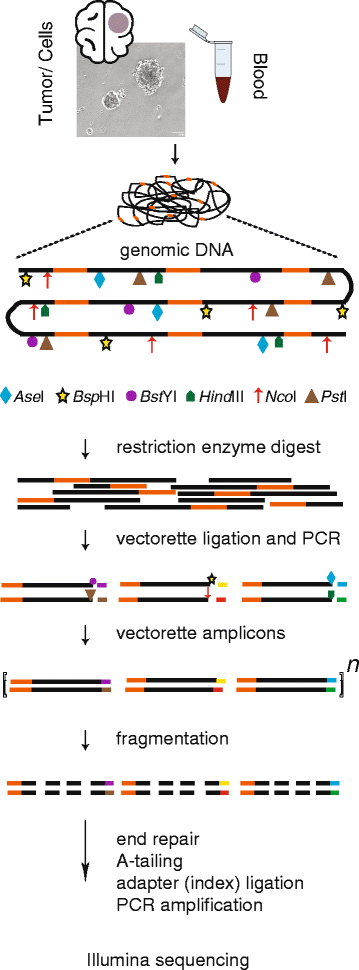
Background: Gliomas are the most common primary brain tumors in adults. We sought to understand the roles of endogenous transposable elements in these malignancies by identifying evidence of somatic retrotransposition in glioblastomas (GBM). We performed transposon insertion profiling of the active subfamily of Long INterspersed Element-1 (LINE-1) elements by deep sequencing (TIPseq) on genomic DNA of low passage oncosphere cell lines derived from 7 primary GBM biopsies, 3 secondary GBM tissue samples, and matched normal intravenous blood samples from the same individuals.
Results: We found and PCR validated one somatically acquired tumor-specific insertion in a case of secondary GBM. No LINE-1 insertions present in primary GBM oncosphere cultures were missing from corresponding blood samples. However, several copies of the element (11) were found in genomic DNA from blood and not in the oncosphere cultures. SNP 6.0 microarray analysis revealed deletions or loss of heterozygosity in the tumor genomes over the intervals corresponding to these LINE-1 insertions.
Conclusions: These findings indicate that LINE-1 retrotransposon can act as an infrequent insertional mutagen in secondary GBM, but that retrotransposition is uncommon in these central nervous system tumors as compared to other neoplasias.
Keywords: Cancer; Glioblastoma; LINE-1; Retrotransposition.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

