A time transect of exomes from a Native American population before and after European contact
- PMID: 27845766
- PMCID: PMC5116069
- DOI: 10.1038/ncomms13175
A time transect of exomes from a Native American population before and after European contact
Abstract
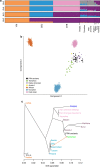
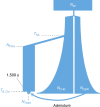
A major factor for the population decline of Native Americans after European contact has been attributed to infectious disease susceptibility. To investigate whether a pre-existing genetic component contributed to this phenomenon, here we analyse 50 exomes of a continuous population from the Northwest Coast of North America, dating from before and after European contact. We model the population collapse after European contact, inferring a 57% reduction in effective population size. We also identify signatures of positive selection on immune-related genes in the ancient but not the modern group, with the strongest signal deriving from the human leucocyte antigen (HLA) gene HLA-DQA1. The modern individuals show a marked frequency decrease in the same alleles, likely due to the environmental change associated with European colonization, whereby negative selection may have acted on the same gene after contact. The evident shift in selection pressures correlates to the regional European-borne epidemics of the 1800s.
Figures
References
-
- Thornton R. Aboriginal North American Population and Rates of Decline, ca. a.d. 1500-1901. Curr. Anthropol. 38, 310–315 (1997).
-
- Thornton R. American Indian Holocaust and Survival University of Oklahoma Press (1987).
-
- Patterson K. B. & Runge T. Smallpox and the Native American. Am. J. Med. Sci. 323, 216 (2002). - PubMed
-
- Boyd R. T. The Coming of the Spirit of Pestilence University of Washington Press (1999).
-
- Dobyns H. F. Disease transfer at contact. Annu. Rev. Anthropol. 22, 273–291 (1993).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

