A Druggable TCF4- and BRD4-Dependent Transcriptional Network Sustains Malignancy in Blastic Plasmacytoid Dendritic Cell Neoplasm
- PMID: 27846392
- PMCID: PMC5175469
- DOI: 10.1016/j.ccell.2016.10.002
A Druggable TCF4- and BRD4-Dependent Transcriptional Network Sustains Malignancy in Blastic Plasmacytoid Dendritic Cell Neoplasm
Abstract
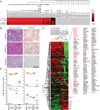
Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is an aggressive and largely incurable hematologic malignancy originating from plasmacytoid dendritic cells (pDCs). Using RNAi screening, we identified the E-box transcription factor TCF4 as a master regulator of the BPDCN oncogenic program. TCF4 served as a faithful diagnostic marker of BPDCN, and its downregulation caused the loss of the BPDCN-specific gene expression program and apoptosis. High-throughput drug screening revealed that bromodomain and extra-terminal domain inhibitors (BETis) induced BPDCN apoptosis, which was attributable to disruption of a BPDCN-specific transcriptional network controlled by TCF4-dependent super-enhancers. BETis retarded the growth of BPDCN xenografts, supporting their clinical evaluation in this recalcitrant malignancy.
Keywords: BPDCN; BRD4; HTS; TCF4; super-enhancer.
Published by Elsevier Inc.
Figures

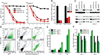





Comment in
-
An Unexpected Chink in the Transcriptional Armor of Plasmacytoid Dendritic Neoplasms.Cancer Cell. 2016 Nov 14;30(5):659-660. doi: 10.1016/j.ccell.2016.10.016. Cancer Cell. 2016. PMID: 27846385
References
-
- Alayed K, Patel KP, Konoplev S, Singh RR, Routbort MJ, Reddy N, Pemmaraju N, Zhang L, Shaikh AA, Aladily TN, et al. TET2 mutations, myelodysplastic features, and a distinct immunoprofile characterize blastic plasmacytoid dendritic cell neoplasm in the bone marrow. American journal of hematology. 2013;88:1055–1061. - PubMed
-
- Assaf C, Gellrich S, Whittaker S, Robson A, Cerroni L, Massone C, Kerl H, Rose C, Chott A, Chimenti S, et al. CD56-positive haematological neoplasms of the skin: a multicentre study of the Cutaneous Lymphoma Project Group of the European Organisation for Research and Treatment of Cancer. J Clin Pathol. 2007;60:981–989. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

