A framework for discrete stochastic simulation on 3D moving boundary domains
- PMID: 27846706
- PMCID: PMC5116030
- DOI: 10.1063/1.4967338
A framework for discrete stochastic simulation on 3D moving boundary domains
Abstract
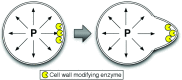
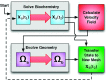
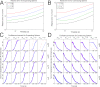
We have developed a method for modeling spatial stochastic biochemical reactions in complex, three-dimensional, and time-dependent domains using the reaction-diffusion master equation formalism. In particular, we look to address the fully coupled problems that arise in systems biology where the shape and mechanical properties of a cell are determined by the state of the biochemistry and vice versa. To validate our method and characterize the error involved, we compare our results for a carefully constructed test problem to those of a microscale implementation. We demonstrate the effectiveness of our method by simulating a model of polarization and shmoo formation during the mating of yeast. The method is generally applicable to problems in systems biology where biochemistry and mechanics are coupled, and spatial stochastic effects are critical.
Figures





Similar articles
-
Modelling of Yeast Mating Reveals Robustness Strategies for Cell-Cell Interactions.PLoS Comput Biol. 2016 Jul 12;12(7):e1004988. doi: 10.1371/journal.pcbi.1004988. eCollection 2016 Jul. PLoS Comput Biol. 2016. PMID: 27404800 Free PMC article.
-
A master equation and moment approach for biochemical systems with creation-time-dependent bimolecular rate functions.J Chem Phys. 2014 Dec 7;141(21):214108. doi: 10.1063/1.4902239. J Chem Phys. 2014. PMID: 25481130 Free PMC article.
-
Particle-based simulations of polarity establishment reveal stochastic promotion of Turing pattern formation.PLoS Comput Biol. 2018 Mar 12;14(3):e1006016. doi: 10.1371/journal.pcbi.1006016. eCollection 2018 Mar. PLoS Comput Biol. 2018. PMID: 29529021 Free PMC article.
-
Stochastic approaches in systems biology.Wiley Interdiscip Rev Syst Biol Med. 2010 Jul-Aug;2(4):385-397. doi: 10.1002/wsbm.78. Wiley Interdiscip Rev Syst Biol Med. 2010. PMID: 20836037 Review.
-
Spatial Stochastic Intracellular Kinetics: A Review of Modelling Approaches.Bull Math Biol. 2019 Aug;81(8):2960-3009. doi: 10.1007/s11538-018-0443-1. Epub 2018 May 21. Bull Math Biol. 2019. PMID: 29785521 Free PMC article. Review.
Cited by
-
Cell polarisation in a bulk-surface model can be driven by both classic and non-classic Turing instability.NPJ Syst Biol Appl. 2021 Feb 26;7(1):13. doi: 10.1038/s41540-021-00173-x. NPJ Syst Biol Appl. 2021. PMID: 33637746 Free PMC article.
-
NERDSS: A Nonequilibrium Simulator for Multibody Self-Assembly at the Cellular Scale.Biophys J. 2020 Jun 16;118(12):3026-3040. doi: 10.1016/j.bpj.2020.05.002. Epub 2020 May 16. Biophys J. 2020. PMID: 32470324 Free PMC article.
-
Reaction rates for reaction-diffusion kinetics on unstructured meshes.J Chem Phys. 2017 Feb 14;146(6):064101. doi: 10.1063/1.4975167. J Chem Phys. 2017. PMID: 28201913 Free PMC article.
-
Three-dimensional stochastic simulation of chemoattractant-mediated excitability in cells.PLoS Comput Biol. 2021 Jul 14;17(7):e1008803. doi: 10.1371/journal.pcbi.1008803. eCollection 2021 Jul. PLoS Comput Biol. 2021. PMID: 34260581 Free PMC article.
-
A hybrid smoothed dissipative particle dynamics (SDPD) spatial stochastic simulation algorithm (sSSA) for advection-diffusion-reaction problems.J Comput Phys. 2019 Feb 1;378:1-17. doi: 10.1016/j.jcp.2018.10.043. Epub 2018 Nov 5. J Comput Phys. 2019. PMID: 31031417 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

