Ribosome biogenesis is dynamically regulated during osteoblast differentiation
- PMID: 27847259
- PMCID: PMC5382099
- DOI: 10.1016/j.gene.2016.11.010
Ribosome biogenesis is dynamically regulated during osteoblast differentiation
Abstract
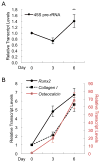
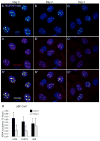
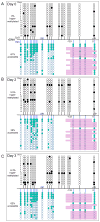
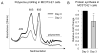
Changes in ribosome biogenesis are tightly linked to cell growth, proliferation, and differentiation. The rate of ribosome biogenesis is established by RNA Pol I-mediated transcription of ribosomal RNA (rRNA). Thus, rRNA gene transcription is a key determinant of cell behavior. Here, we show that ribosome biogenesis is dynamically regulated during osteoblast differentiation. Upon osteoinduction, osteoprogenitor cells transiently silence a subset of rRNA genes through a reversible mechanism that is initiated through biphasic nucleolar depletion of UBF1 and then RNA Pol I. Nucleolar depletion of UBF1 is coincident with an increase in the number of silent but transcriptionally permissible rRNA genes. This increase in the number of silent rRNA genes reduces levels of ribosome biogenesis and subsequently, protein synthesis. Together these findings demonstrate that fluctuations in rRNA gene transcription are determined by nucleolar occupancy of UBF1 and closely coordinated with the early events necessary for acquisition of the osteoblast cell fate.
Keywords: Bone; Osteoblast; Osteoprogenitor; RNA Pol I; Ribosome biogenesis; UBF1; rRNA.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

