The roles of RNA processing in translating genotype to phenotype
- PMID: 27847391
- PMCID: PMC5544131
- DOI: 10.1038/nrm.2016.139
The roles of RNA processing in translating genotype to phenotype
Abstract
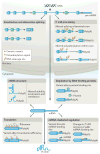
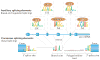
A goal of human genetics studies is to determine the mechanisms by which genetic variation produces phenotypic differences that affect human health. Efforts in this respect have previously focused on genetic variants that affect mRNA levels by altering epigenetic and transcriptional regulation. Recent studies show that genetic variants that affect RNA processing are at least equally as common as, and are largely independent from, those variants that affect transcription. We highlight the impact of genetic variation on pre-mRNA splicing and polyadenylation, and on the stability, translation and structure of mRNAs as mechanisms that produce phenotypic traits. These results emphasize the importance of including RNA processing signals in analyses to identify functional variants.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Kwan T, et al. Genome-wide analysis of transcript isoform variation in humans. Nat Genet. 2008;40:225–231. One of the early genome-wide comparisons showed that genomic variation between individuals correlated with differences in mRNA levels and structure. - PubMed
-
- Li YI, et al. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. A systematic analysis of LCLs from 68 individuals identified genomic variants that correlate with differences in up to eight molecular features of the gene regulatory cascade. - PMC - PubMed
-
- Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. This study identified the genomic causes of variation in the transcriptome using RNA-seq and genomic sequence data from LCLs of 462 individuals in the 1000 Genomes Project. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

