Functional expression and evaluation of heterologous phosphoketolases in Saccharomyces cerevisiae
- PMID: 27848233
- PMCID: PMC5110461
- DOI: 10.1186/s13568-016-0290-0
Functional expression and evaluation of heterologous phosphoketolases in Saccharomyces cerevisiae
Abstract
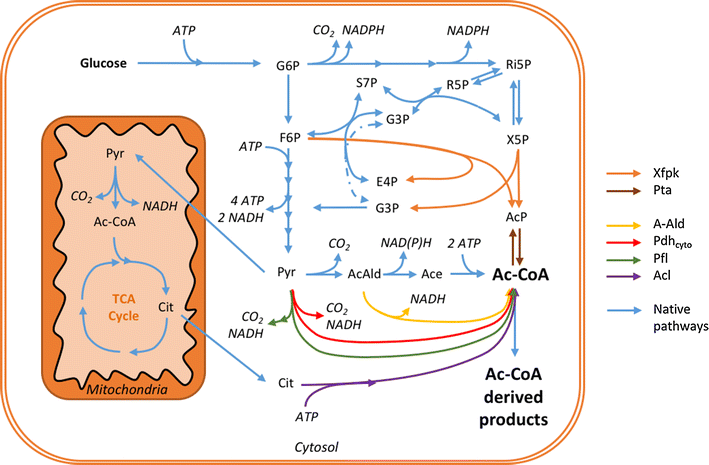
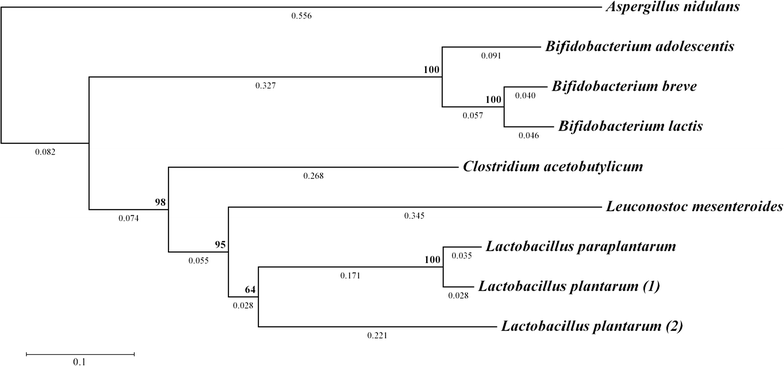
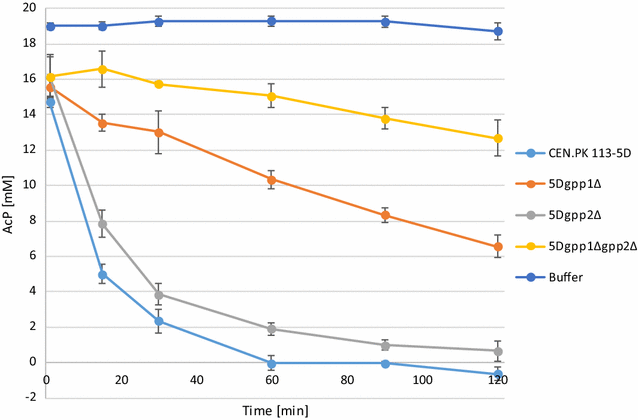
Phosphoketolases catalyze an energy- and redox-independent cleavage of certain sugar phosphates. Hereby, the two-carbon (C2) compound acetyl-phosphate is formed, which enzymatically can be converted into acetyl-CoA-a key precursor in central carbon metabolism. Saccharomyces cerevisiae does not demonstrate efficient phosphoketolase activity naturally. In this study, we aimed to compare and identify efficient heterologous phosphoketolase enzyme candidates that in yeast have the potential to reduce carbon loss compared to the native acetyl-CoA producing pathway by redirecting carbon flux directly from C5 and C6 sugars towards C2-synthesis. Nine phosphoketolase candidates were expressed in S. cerevisiae of which seven produced significant amounts of acetyl-phosphate after provision of sugar phosphate substrates in vitro. The candidates showed differing substrate specificities, and some demonstrated activity levels significantly exceeding those of candidates previously expressed in yeast. The conducted studies also revealed that S. cerevisiae contains endogenous enzymes capable of breaking down acetyl-phosphate, likely into acetate, and that removal of the phosphatases Gpp1 and Gpp2 could largely prevent this breakdown. An evaluation of in vivo function of a subset of phosphoketolases was conducted by monitoring acetate levels during growth, confirming that candidates showing high activity in vitro indeed showed increased acetate accumulation, but expression also decreased cellular fitness. The study shows that expression of several bacterial phosphoketolase candidates in S. cerevisiae can efficiently divert intracellular carbon flux toward C2-synthesis, thus showing potential to be used in metabolic engineering strategies aimed to increase yields of acetyl-CoA derived compounds.
Keywords: Acetyl-CoA; Acetyl-phosphate; Carbon efficiency; Phosphoketolase; Saccharomyces cerevisiae.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

