17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells
- PMID: 27848966
- PMCID: PMC5116091
- DOI: 10.1038/ncomms13426
17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells
Abstract
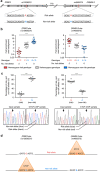
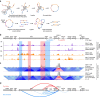
Asthma and autoimmune disease susceptibility has been strongly linked to genetic variants in the 17q21 haploblock that alter the expression of ORMDL3; however, the molecular mechanisms by which these variants perturb gene expression and the cell types in which this effect is most prominent are unclear. We found several 17q21 variants overlapped enhancers present mainly in primary immune cell types. CD4+ T cells showed the greatest increase (threefold) in ORMDL3 expression in individuals carrying the asthma-risk alleles, where ORMDL3 negatively regulated interleukin-2 production. The asthma-risk variants rs4065275 and rs12936231 switched CTCF-binding sites in the 17q21 locus, and 4C-Seq assays showed that several distal cis-regulatory elements upstream of the disrupted ZPBP2 CTCF-binding site interacted with the ORMDL3 promoter region in CD4+ T cells exclusively from subjects carrying asthma-risk alleles. Overall, our results suggested that T cells are one of the most prominent cell types affected by 17q21 variants.
Figures




References
-
- Moffatt M. F. et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448, 470–U475 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

