Epigenetic and genetic components of height regulation
- PMID: 27848971
- PMCID: PMC5116096
- DOI: 10.1038/ncomms13490
Epigenetic and genetic components of height regulation
Abstract
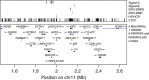
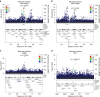
Adult height is a highly heritable trait. Here we identified 31.6 million sequence variants by whole-genome sequencing of 8,453 Icelanders and tested them for association with adult height by imputing them into 88,835 Icelanders. Here we discovered 13 novel height associations by testing four different models including parent-of-origin (|β|=0.4-10.6 cm). The minor alleles of three parent-of-origin signals associate with less height only when inherited from the father and are located within imprinted regions (IGF2-H19 and DLK1-MEG3). We also examined the association of these sequence variants in a set of 12,645 Icelanders with birth length measurements. Two of the novel variants, (IGF2-H19 and TET1), show significant association with both adult height and birth length, indicating a role in early growth regulation. Among the parent-of-origin signals, we observed opposing parental effects raising questions about underlying mechanisms. These findings demonstrate that common variations affect human growth by parental imprinting.
Conflict of interest statement
The following authors affiliated with deCODE genetics/Amgen are employed by the company: S.B., A.O., A.H., R.P.K., G.S., A.O., G.T., O.B.D., G.A.A, G.S., B.O.J., H.H., K.F.A., G.B.W., S.A.G., P.D.I. L.D.W., J.K.S., M.L.F., T.R., A.K., G.M., H. Helgason, U.T., D.F.G., P.S. and K.S.
Figures

References
-
- Silventoinen K. et al. Heritability of adult body height: a comparative study of twin cohorts in eight countries. Twin Res. 6, 399–408 (2003). - PubMed
-
- Carmichael C. M. & McGue M. A cross-sectional examination of height, weight, and body mass index in adult twins. J. Gerontol. A. Biol. Sci. Med. Sci. 50, B237–B244 (1995). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

