Contribution of Asparagine Catabolism to Salmonella Virulence
- PMID: 27849183
- PMCID: PMC5278173
- DOI: 10.1128/IAI.00740-16
Contribution of Asparagine Catabolism to Salmonella Virulence
Abstract
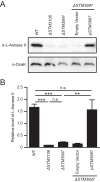
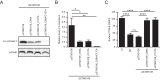
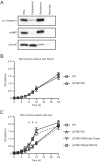
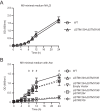
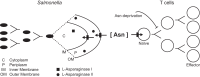
Salmonellae are pathogenic bacteria that cause significant morbidity and mortality in humans worldwide. Salmonellae establish infection and avoid clearance by the immune system by mechanisms that are not well understood. We previously showed that l-asparaginase II produced by Salmonella enterica serovar Typhimurium (S Typhimurium) inhibits T cell responses and mediates virulence. In addition, we previously showed that asparagine deprivation such as that mediated by l-asparaginase II of S Typhimurium causes suppression of activation-induced T cell metabolic reprogramming. Here, we report that STM3997, which encodes a homolog of disulfide bond protein A (dsbA) of Escherichia coli, is required for l-asparaginase II stability and function. Furthermore, we report that l-asparaginase II localizes primarily to the periplasm and acts together with l-asparaginase I to provide S Typhimurium the ability to catabolize asparagine and assimilate nitrogen. Importantly, we determined that, in a murine model of infection, S Typhimurium lacking both l-asparaginase I and II genes competes poorly with wild-type S Typhimurium for colonization of target tissues. Collectively, these results indicate that asparagine catabolism contributes to S Typhimurium virulence, providing new insights into the competition for nutrients at the host-pathogen interface.
Keywords: Salmonella; T cells; asparaginase; asparagine; catabolism; host response; metabolism; nitrogen metabolism; pathogenesis; virulence.
Copyright © 2017 American Society for Microbiology.
Figures

References
-
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness” Studies . 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi:10.1086/650733. - DOI - PubMed
-
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Dopfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001940. doi:10.1371/journal.pmed.1001940. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

