Origins of the current seventh cholera pandemic
- PMID: 27849586
- PMCID: PMC5137724
- DOI: 10.1073/pnas.1608732113
Origins of the current seventh cholera pandemic
Abstract
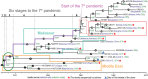
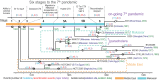
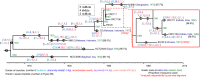
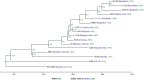
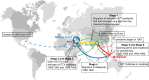
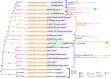
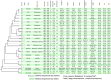
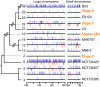
Vibrio cholerae has caused seven cholera pandemics since 1817, imposing terror on much of the world, but bacterial strains are currently only available for the sixth and seventh pandemics. The El Tor biotype seventh pandemic began in 1961 in Indonesia, but did not originate directly from the classical biotype sixth-pandemic strain. Previous studies focused mainly on the spread of the seventh pandemic after 1970. Here, we analyze in unprecedented detail the origin, evolution, and transition to pandemicity of the seventh-pandemic strain. We used high-resolution comparative genomic analysis of strains collected from 1930 to 1964, covering the evolution from the first available El Tor biotype strain to the start of the seventh pandemic. We define six stages leading to the pandemic strain and reveal all key events. The seventh pandemic originated from a nonpathogenic strain in the Middle East, first observed in 1897. It subsequently underwent explosive diversification, including the spawning of the pandemic lineage. This rapid diversification suggests that, when first observed, the strain had only recently arrived in the Middle East, possibly from the Asian homeland of cholera. The lineage migrated to Makassar, Indonesia, where it gained the important virulence-associated elements Vibrio seventh pandemic island I (VSP-I), VSP-II, and El Tor type cholera toxin prophage by 1954, and it then became pandemic in 1961 after only 12 additional mutations. Our data indicate that specific niches in the Middle East and Makassar were important in generating the pandemic strain by providing gene sources and the driving forces for genetic events.
Keywords: Vibrio cholerae; comparative genomics; evolution; pandemic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Barua D. History of cholera. In: Barua D, Greenough WB III, editors. Cholera. Plenum; New York: 1992. pp. 1–36.
-
- Pan American Health Organization and World Health Organization . Epidemiological Update - Cholera - 18 February 2014. Pan American Health Organization/World Health Organization; Washington, DC: 2014.
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

