Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis
- PMID: 27849615
- PMCID: PMC5137743
- DOI: 10.1073/pnas.1612635113
Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis
Abstract
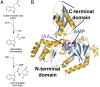
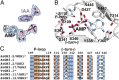
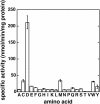
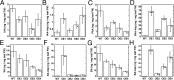
In Arabidopsis thaliana, the acyl acid amido synthetase Gretchen Hagen 3.5 (AtGH3.5) conjugates both indole-3-acetic acid (IAA) and salicylic acid (SA) to modulate auxin and pathogen response pathways. To understand the molecular basis for the activity of AtGH3.5, we determined the X-ray crystal structure of the enzyme in complex with IAA and AMP. Biochemical analysis demonstrates that the substrate preference of AtGH3.5 is wider than originally described and includes the natural auxin phenylacetic acid (PAA) and the potential SA precursor benzoic acid (BA). Residues that determine IAA versus BA substrate preference were identified. The dual functionality of AtGH3.5 is unique to this enzyme although multiple IAA-conjugating GH3 proteins share nearly identical acyl acid binding sites. In planta analysis of IAA, PAA, SA, and BA and their respective aspartyl conjugates were determined in wild-type and overexpressing lines of A thaliana This study suggests that AtGH3.5 conjugates auxins (i.e., IAA and PAA) and benzoates (i.e., SA and BA) to mediate crosstalk between different metabolic pathways, broadening the potential roles for GH3 acyl acid amido synthetases in plants.
Keywords: Arabidopsis; auxin; plant biochemistry; plant hormone; protein structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Staswick PE, Tiryaki I, Rowe ML. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14(6):1405–1415. - PMC - PubMed
-
- Fonseca S, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5(5):344–350. - PubMed
-
- Meesters C, et al. A chemical inhibitor of jasmonate signaling targets JAR1 in Arabidopsis thaliana. Nat Chem Biol. 2014;10(10):830–836. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

