Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair
- PMID: 27851738
- PMCID: PMC5845140
- DOI: 10.1038/nature20562
Cascading MutS and MutL sliding clamps control DNA diffusion to activate mismatch repair
Abstract
Mismatched nucleotides arise from polymerase misincorporation errors, recombination between heteroallelic parents and chemical or physical DNA damage. Highly conserved MutS (MSH) and MutL (MLH/PMS) homologues initiate mismatch repair and, in higher eukaryotes, act as DNA damage sensors that can trigger apoptosis. Defects in human mismatch repair genes cause Lynch syndrome or hereditary non-polyposis colorectal cancer and 10-40% of related sporadic tumours. However, the collaborative mechanics of MSH and MLH/PMS proteins have not been resolved in any organism. We visualized Escherichia coli (Ec) ensemble mismatch repair and confirmed that EcMutS mismatch recognition results in the formation of stable ATP-bound sliding clamps that randomly diffuse along the DNA with intermittent backbone contact. The EcMutS sliding clamps act as a platform to recruit EcMutL onto the mismatched DNA, forming an EcMutS-EcMutL search complex that then closely follows the DNA backbone. ATP binding by EcMutL establishes a second long-lived DNA clamp that oscillates between the principal EcMutS-EcMutL search complex and unrestricted EcMutS and EcMutL sliding clamps. The EcMutH endonuclease that targets mismatch repair excision only binds clamped EcMutL, increasing its DNA association kinetics by more than 1,000-fold. The assembly of an EcMutS-EcMutL-EcMutH search complex illustrates how sequential stable sliding clamps can modulate one-dimensional diffusion mechanics along the DNA to direct mismatch repair.
Figures


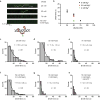


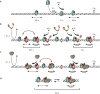
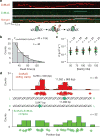



Comment in
-
DNA repair: Clamping down on copy errors.Nature. 2016 Nov 24;539(7630):498-499. doi: 10.1038/nature20475. Epub 2016 Nov 16. Nature. 2016. PMID: 27851733 Free PMC article.
Similar articles
-
MutL sliding clamps coordinate exonuclease-independent Escherichia coli mismatch repair.Nat Commun. 2019 Nov 22;10(1):5294. doi: 10.1038/s41467-019-13191-5. Nat Commun. 2019. PMID: 31757945 Free PMC article.
-
MutS functions as a clamp loader by positioning MutL on the DNA during mismatch repair.Nat Commun. 2022 Oct 3;13(1):5808. doi: 10.1038/s41467-022-33479-3. Nat Commun. 2022. PMID: 36192430 Free PMC article.
-
MutS homolog sliding clamps shield the DNA from binding proteins.J Biol Chem. 2018 Sep 14;293(37):14285-14294. doi: 10.1074/jbc.RA118.002264. Epub 2018 Aug 2. J Biol Chem. 2018. PMID: 30072380 Free PMC article.
-
Mismatch repair.J Biol Chem. 2015 Oct 30;290(44):26395-403. doi: 10.1074/jbc.R115.660142. Epub 2015 Sep 9. J Biol Chem. 2015. PMID: 26354434 Free PMC article. Review.
-
Stochastic Processes and Component Plasticity Governing DNA Mismatch Repair.J Mol Biol. 2018 Oct 26;430(22):4456-4468. doi: 10.1016/j.jmb.2018.05.039. Epub 2018 Jun 1. J Mol Biol. 2018. PMID: 29864444 Free PMC article. Review.
Cited by
-
Coarse-grained molecular dynamics simulations of base-pair mismatch recognition protein MutS sliding along DNA.Biophys Physicobiol. 2022 Apr 14;19:1-16. doi: 10.2142/biophysico.bppb-v19.0015. eCollection 2022. Biophys Physicobiol. 2022. PMID: 35797408 Free PMC article.
-
Endonuclease-independent DNA mismatch repair processes on the lagging strand.DNA Repair (Amst). 2018 Aug;68:41-49. doi: 10.1016/j.dnarep.2018.06.002. Epub 2018 Jun 12. DNA Repair (Amst). 2018. PMID: 29929046 Free PMC article.
-
Handcuffing intrinsically disordered regions in Mlh1-Pms1 disrupts mismatch repair.Nucleic Acids Res. 2021 Sep 20;49(16):9327-9341. doi: 10.1093/nar/gkab694. Nucleic Acids Res. 2021. PMID: 34390347 Free PMC article.
-
Glioblastoma multiforme: an updated overview of temozolomide resistance mechanisms and strategies to overcome resistance.Discov Oncol. 2025 May 12;16(1):731. doi: 10.1007/s12672-025-02567-3. Discov Oncol. 2025. PMID: 40353925 Free PMC article. Review.
-
Evolutionary advantage of a dissociative search mechanism in DNA mismatch repair.Phys Rev E. 2021 May;103(5-1):052404. doi: 10.1103/PhysRevE.103.052404. Phys Rev E. 2021. PMID: 34134264 Free PMC article.
References
-
- Wang JY, Edelmann W. Mismatch repair proteins as sensors of alkylation DNA damage. Cancer Cell. 2006;9:417–418. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

