Variant Exported Blood-Stage Proteins Encoded by Plasmodium Multigene Families Are Expressed in Liver Stages Where They Are Exported into the Parasitophorous Vacuole
- PMID: 27851824
- PMCID: PMC5113031
- DOI: 10.1371/journal.ppat.1005917
Variant Exported Blood-Stage Proteins Encoded by Plasmodium Multigene Families Are Expressed in Liver Stages Where They Are Exported into the Parasitophorous Vacuole
Erratum in
-
Correction: Variant Exported Blood-Stage Proteins Encoded by Plasmodium Multigene Families Are Expressed in Liver Stages Where They Are Exported into the Parasitophorous Vacuole.PLoS Pathog. 2016 Dec 14;12(12):e1006107. doi: 10.1371/journal.ppat.1006107. eCollection 2016 Dec. PLoS Pathog. 2016. PMID: 27973555 Free PMC article.
-
Correction: Variant Exported Blood-Stage Proteins Encoded by Plasmodium Multigene Families Are Expressed in Liver Stages Where They Are Exported into the Parasitophorous Vacuole.PLoS Pathog. 2017 Jan 17;13(1):e1006128. doi: 10.1371/journal.ppat.1006128. eCollection 2017 Jan. PLoS Pathog. 2017. PMID: 28095481 Free PMC article.
Abstract
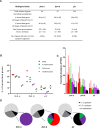
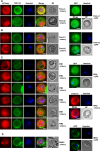
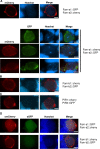
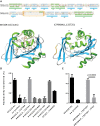
Many variant proteins encoded by Plasmodium-specific multigene families are exported into red blood cells (RBC). P. falciparum-specific variant proteins encoded by the var, stevor and rifin multigene families are exported onto the surface of infected red blood cells (iRBC) and mediate interactions between iRBC and host cells resulting in tissue sequestration and rosetting. However, the precise function of most other Plasmodium multigene families encoding exported proteins is unknown. To understand the role of RBC-exported proteins of rodent malaria parasites (RMP) we analysed the expression and cellular location by fluorescent-tagging of members of the pir, fam-a and fam-b multigene families. Furthermore, we performed phylogenetic analyses of the fam-a and fam-b multigene families, which indicate that both families have a history of functional differentiation unique to RMP. We demonstrate for all three families that expression of family members in iRBC is not mutually exclusive. Most tagged proteins were transported into the iRBC cytoplasm but not onto the iRBC plasma membrane, indicating that they are unlikely to play a direct role in iRBC-host cell interactions. Unexpectedly, most family members are also expressed during the liver stage, where they are transported into the parasitophorous vacuole. This suggests that these protein families promote parasite development in both the liver and blood, either by supporting parasite development within hepatocytes and erythrocytes and/or by manipulating the host immune response. Indeed, in the case of Fam-A, which have a steroidogenic acute regulatory-related lipid transfer (START) domain, we found that several family members can transfer phosphatidylcholine in vitro. These observations indicate that these proteins may transport (host) phosphatidylcholine for membrane synthesis. This is the first demonstration of a biological function of any exported variant protein family of rodent malaria parasites.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Proteomic and genetic analyses demonstrate that Plasmodium berghei blood stages export a large and diverse repertoire of proteins.Mol Cell Proteomics. 2013 Feb;12(2):426-48. doi: 10.1074/mcp.M112.021238. Epub 2012 Nov 28. Mol Cell Proteomics. 2013. PMID: 23197789 Free PMC article.
-
The Plasmodium falciparum STEVOR multigene family mediates antigenic variation of the infected erythrocyte.PLoS Pathog. 2009 Feb;5(2):e1000307. doi: 10.1371/journal.ppat.1000307. Epub 2009 Feb 20. PLoS Pathog. 2009. PMID: 19229319 Free PMC article.
-
Differential Trafficking and Expression of PIR Proteins in Acute and Chronic Plasmodium Infections.Front Cell Infect Microbiol. 2022 Jun 16;12:877253. doi: 10.3389/fcimb.2022.877253. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35782145 Free PMC article.
-
Variable Surface Antigens of Plasmodium falciparum: Protein Families with Divergent Roles.Protein Pept Lett. 2024;31(6):409-423. doi: 10.2174/0109298665298567240530170924. Protein Pept Lett. 2024. PMID: 38910420 Review.
-
Variant surface antigens of Plasmodium falciparum and their roles in severe malaria.Nat Rev Microbiol. 2017 Aug;15(8):479-491. doi: 10.1038/nrmicro.2017.47. Epub 2017 Jun 12. Nat Rev Microbiol. 2017. PMID: 28603279 Review.
Cited by
-
FT-GPI, a highly sensitive and accurate predictor of GPI-anchored proteins, reveals the composition and evolution of the GPI proteome in Plasmodium species.Malar J. 2023 Jan 25;22(1):27. doi: 10.1186/s12936-022-04430-0. Malar J. 2023. PMID: 36698187 Free PMC article.
-
Sexual differentiation in human malaria parasites is regulated by competition between phospholipid metabolism and histone methylation.Nat Microbiol. 2023 Jul;8(7):1280-1292. doi: 10.1038/s41564-023-01396-w. Epub 2023 Jun 5. Nat Microbiol. 2023. PMID: 37277533 Free PMC article.
-
Transcriptional heterogeneity and tightly regulated changes in gene expression during Plasmodium berghei sporozoite development.Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2023438118. doi: 10.1073/pnas.2023438118. Proc Natl Acad Sci U S A. 2021. PMID: 33653959 Free PMC article.
-
Host cell cytosolic immune response during Plasmodium liver stage development.FEMS Microbiol Rev. 2018 May 1;42(3):324-334. doi: 10.1093/femsre/fuy007. FEMS Microbiol Rev. 2018. PMID: 29529207 Free PMC article. Review.
-
Correction: Variant Exported Blood-Stage Proteins Encoded by Plasmodium Multigene Families Are Expressed in Liver Stages Where They Are Exported into the Parasitophorous Vacuole.PLoS Pathog. 2017 Jan 17;13(1):e1006128. doi: 10.1371/journal.ppat.1006128. eCollection 2017 Jan. PLoS Pathog. 2017. PMID: 28095481 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

