Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer
- PMID: 27851913
- PMCID: PMC5134901
- DOI: 10.1016/j.immuni.2016.10.032
Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer
Abstract
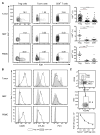
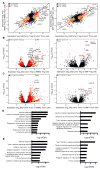
Regulatory T (Treg) cells reside in lymphoid organs and barrier tissues where they control different types of inflammatory responses. Treg cells are also found in human cancers, and studies in animal models suggest that they contribute to cancer progression. However, properties of human intratumoral Treg cells and those present in corresponding normal tissue remain largely unknown. Here, we analyzed features of Treg cells in untreated human breast carcinomas, normal mammary gland, and peripheral blood. Tumor-resident Treg cells were potently suppressive and their gene-expression pattern resembled that of normal breast tissue, but not of activated peripheral blood Treg cells. Nevertheless, a number of cytokine and chemokine receptor genes, most notably CCR8, were upregulated in tumor-resident Treg cells in comparison to normal tissue-resident ones. Our studies suggest that targeting CCR8 for the depletion of tumor-resident Treg cells might represent a promising immunotherapeutic approach for the treatment of breast cancer.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures





Comment in
-
Tumour immunology: Interrogating intratumoral Treg cells.Nat Rev Immunol. 2017 Jan;17(1):4-5. doi: 10.1038/nri.2016.137. Epub 2016 Dec 5. Nat Rev Immunol. 2017. PMID: 27916981 No abstract available.
References
-
- Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL, Banham AH. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:5373–5380. - PubMed
-
- Bohling SD, Allison KH. Immunosuppressive regulatory T cells are associated with aggressive breast cancer phenotypes: a potential therapeutic target. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;21:1527–1532. - PubMed
-
- Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, Chudakov DM. MiXCR: software for comprehensive adaptive immunity profiling. Nature methods. 2015;12:380–381. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

