Modeling and Simulating the Dynamics of Type IV Pili Extension of Pseudomonas aeruginosa
- PMID: 27851948
- PMCID: PMC5112937
- DOI: 10.1016/j.bpj.2016.09.050
Modeling and Simulating the Dynamics of Type IV Pili Extension of Pseudomonas aeruginosa
Abstract
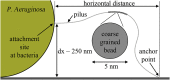
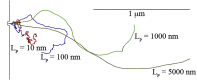
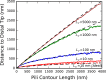
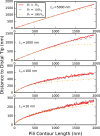
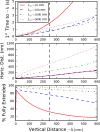
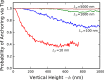
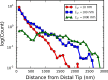
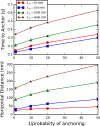
Bacteria such as Pseudomonas aeruginosa use type IV pili to move across surfaces. The pili extend, attach to the surface, and then retract to move the bacteria forward. In this article, a coarse-grained model of pilus extension and attachment is developed. Simulations performed at biologically relevant conditions indicate that pilus extension is a quasistatic process such that the pili are able to relax via thermal fluctuations as it is being built and extended. Results are generated for pili with different rigidities ranging from very flexible to very stiff. It is shown that very flexible pili do not extend very far and thus would limit the bacteria to short jumps forward while stiff pili enable much greater displacements. Feasible mechanisms of attachment to the surface are also found to vary greatly between flexible and stiff pili. While it is not always the tip of flexible pili that first makes contact with the substrate, it is likely to be a part of the pili that is close to the tip. Conversely, stiff pili are much more likely to make contact with the substrate via the tip, but if not then the part of the pilus that attaches can be quite far from the tip. These results thus give insight to help resolve current discrepancies in the literature regarding pilus stiffness and the location of adhesins on pili.
Copyright © 2016 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
The binding of Pseudomonas aeruginosa pili to glycosphingolipids is a tip-associated event involving the C-terminal region of the structural pilin subunit.Mol Microbiol. 1994 Feb;11(4):705-13. doi: 10.1111/j.1365-2958.1994.tb00348.x. Mol Microbiol. 1994. PMID: 7910938
-
DNA binding: a novel function of Pseudomonas aeruginosa type IV pili.J Bacteriol. 2005 Feb;187(4):1455-64. doi: 10.1128/JB.187.4.1455-1464.2005. J Bacteriol. 2005. PMID: 15687210 Free PMC article.
-
Direct observation of extension and retraction of type IV pili.Proc Natl Acad Sci U S A. 2001 Jun 5;98(12):6901-4. doi: 10.1073/pnas.121171698. Epub 2001 May 29. Proc Natl Acad Sci U S A. 2001. PMID: 11381130 Free PMC article.
-
Chaperone-assisted assembly and molecular architecture of adhesive pili.Annu Rev Microbiol. 1991;45:383-415. doi: 10.1146/annurev.mi.45.100191.002123. Annu Rev Microbiol. 1991. PMID: 1683764 Review.
-
Pili with strong attachments: Gram-positive bacteria do it differently.Mol Microbiol. 2006 Oct;62(2):320-30. doi: 10.1111/j.1365-2958.2006.05279.x. Epub 2006 Sep 15. Mol Microbiol. 2006. PMID: 16978260 Review.
Cited by
-
Modelling bacterial twitching in fluid flows: a CFD-DEM approach.Sci Rep. 2019 Oct 10;9(1):14540. doi: 10.1038/s41598-019-51101-3. Sci Rep. 2019. PMID: 31601892 Free PMC article.
-
Data-driven modelling makes quantitative predictions regarding bacteria surface motility.PLoS Comput Biol. 2024 May 14;20(5):e1012063. doi: 10.1371/journal.pcbi.1012063. eCollection 2024 May. PLoS Comput Biol. 2024. PMID: 38743804 Free PMC article.
-
Fresh Extension of Vibrio cholerae Competence Type IV Pili Predisposes Them for Motor-Independent Retraction.Appl Environ Microbiol. 2021 Jun 25;87(14):e0047821. doi: 10.1128/AEM.00478-21. Epub 2021 Jun 25. Appl Environ Microbiol. 2021. PMID: 33990308 Free PMC article.
References
-
- Bradley D.E. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 1980;26:146–154. - PubMed
-
- Semmler A.B., Whitchurch C.B., Mattick J.S. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology. 1999;145:2863–2873. - PubMed
-
- Merz A.J., So M., Sheetz M.P. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

