Conserved Tetramer Junction in the Kinetochore Ndc80 Complex
- PMID: 27851957
- PMCID: PMC5131873
- DOI: 10.1016/j.celrep.2016.10.065
Conserved Tetramer Junction in the Kinetochore Ndc80 Complex
Abstract
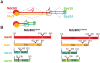
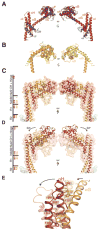
The heterotetrameric Ndc80 complex establishes connectivity along the principal longitudinal axis of a kinetochore. Its two heterodimeric subcomplexes, each with a globular end and a coiled-coil shaft, connect end-to-end to create a ∼600 Å long rod spanning the gap from centromere-proximal structures to spindle microtubules. Neither subcomplex has a known function on its own, but the heterotetrameric organization and the characteristics of the junction are conserved from yeast to man. We have determined crystal structures of two shortened ("dwarf") Ndc80 complexes that contain the full tetramer junction and both globular ends. The junction connects two α-helical coiled coils through regions of four-chain and three-chain overlap. The complexity of its structure depends on interactions among conserved amino-acid residues, suggesting a binding site for additional cellular factor(s) not yet identified.
Keywords: X-ray crystallography; cell division; coiled-coil junction; kinetochore assembly; kinetochore structure.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Molecular organization of the Ndc80 complex, an essential kinetochore component.Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5363-7. doi: 10.1073/pnas.0501168102. Epub 2005 Apr 4. Proc Natl Acad Sci U S A. 2005. PMID: 15809444 Free PMC article.
-
Kinetochore biorientation in Saccharomyces cerevisiae requires a tightly folded conformation of the Ndc80 complex.Genetics. 2014 Dec;198(4):1483-93. doi: 10.1534/genetics.114.167775. Epub 2014 Sep 16. Genetics. 2014. PMID: 25230952 Free PMC article.
-
Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex.J Mol Biol. 2011 Jan 14;405(2):548-59. doi: 10.1016/j.jmb.2010.11.012. Epub 2010 Nov 12. J Mol Biol. 2011. PMID: 21075115 Free PMC article.
-
The Ndc80 complex: hub of kinetochore activity.FEBS Lett. 2007 Jun 19;581(15):2862-9. doi: 10.1016/j.febslet.2007.05.012. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17521635 Review.
-
Structures and functions of yeast kinetochore complexes.Annu Rev Biochem. 2007;76:563-91. doi: 10.1146/annurev.biochem.76.052705.160607. Annu Rev Biochem. 2007. PMID: 17362199 Review.
Cited by
-
A conserved site on Ndc80 complex facilitates dynamic recruitment of Mps1 to yeast kinetochores to promote accurate chromosome segregation.Curr Biol. 2024 Jun 3;34(11):2294-2307.e4. doi: 10.1016/j.cub.2024.04.054. Epub 2024 May 21. Curr Biol. 2024. PMID: 38776906 Free PMC article.
-
Methylation of CENP-A/Cse4 on arginine 143 and lysine 131 regulates kinetochore stability in yeast.Genetics. 2023 Apr 6;223(4):iyad028. doi: 10.1093/genetics/iyad028. Genetics. 2023. PMID: 36810679 Free PMC article.
-
Three interacting regions of the Ndc80 and Dam1 complexes support microtubule tip-coupling under load.J Cell Biol. 2022 May 2;221(5):e202107016. doi: 10.1083/jcb.202107016. Epub 2022 Mar 30. J Cell Biol. 2022. PMID: 35353161 Free PMC article.
-
The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle.Open Biol. 2020 Jun;10(6):200051. doi: 10.1098/rsob.200051. Epub 2020 Jun 10. Open Biol. 2020. PMID: 32516549 Free PMC article. Review.
-
An interaction hub on Ndc80 complex facilitates dynamic recruitment of Mps1 to yeast kinetochores to promote accurate chromosome segregation.bioRxiv [Preprint]. 2023 Nov 7:2023.11.07.566082. doi: 10.1101/2023.11.07.566082. bioRxiv. 2023. Update in: Curr Biol. 2024 Jun 3;34(11):2294-2307.e4. doi: 10.1016/j.cub.2024.04.054. PMID: 37986816 Free PMC article. Updated. Preprint.
References
-
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

