Epigenetic silencing of TET2 and TET3 induces an EMT-like process in melanoma
- PMID: 27852070
- PMCID: PMC5352122
- DOI: 10.18632/oncotarget.13324
Epigenetic silencing of TET2 and TET3 induces an EMT-like process in melanoma
Abstract
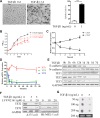
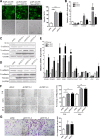
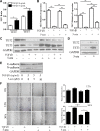
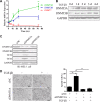
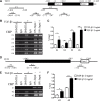
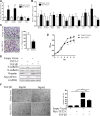
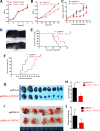
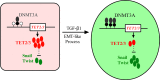
Epithelial-Mesenchymal Transition (EMT) is a critical step in the progression of cancer. Malignant melanoma, a cancer developed from pigmented melanocytes, metastasizes through an EMT-like process. Ten-eleven translocation (TET) enzymes, catalyzing the conversion of 5-methylcytosine (5mC) to 5-hydroxylmethylcytosine (5-hmC), are down regulated in melanoma. However, their roles in the progression and the EMT-like process of melanoma are not fully understood. Here we report that DNA methylation induced silencing of TET2 and TET3 are responsible for the EMT-like process and the metastasis of melanoma. TET2 and TET3 are down regulated in the TGF-β1-induced EMT-like process, and the knocking down of TET2 or TET3 induced this EMT-like process. A DNA demethylating agent antagonized the TGF-β-induced suppression of TET2 and TET3. Furthermore, a ChIP analysis indicated that enhanced recruitment of DNMT3A (DNA Methyltransferase 3A) is the mechanism by which TGF-β induces the silencing of TET2 and TET3. Finally, the overexpression of the TET2 C-terminal sequence partially rescues the TGF-β1-induced EMT-like process in vitro and inhibits tumor growth and metastasis in vivo. Hence, our data suggest an epigenetic circuitry that mediates the EMT activated by TGF-β. As an effector, DNMT3A senses the TGF-β signal and silences TET2 and TET3 promoters to induce the EMT-like process and metastasis in melanoma.
Keywords: DNMT3A; EMT; Melanoma; TET; TGF-β.
Conflict of interest statement
All the authors claim here that there is no potential conflicts of interests.
Figures
Similar articles
-
TET3 inhibits TGF-β1-induced epithelial-mesenchymal transition by demethylating miR-30d precursor gene in ovarian cancer cells.J Exp Clin Cancer Res. 2016 May 4;35:72. doi: 10.1186/s13046-016-0350-y. J Exp Clin Cancer Res. 2016. PMID: 27141829 Free PMC article.
-
DNMT3B attenuated the inhibition of TET3 on epithelial-mesenchymal transition in TGF-β1-induced ovarian cancer by methylating the TET3 promoter.Reprod Biol. 2022 Dec;22(4):100701. doi: 10.1016/j.repbio.2022.100701. Epub 2022 Oct 12. Reprod Biol. 2022. PMID: 36242939
-
TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS.EMBO J. 2013 Mar 6;32(5):645-55. doi: 10.1038/emboj.2012.357. Epub 2013 Jan 25. EMBO J. 2013. PMID: 23353889 Free PMC article.
-
TETology: Epigenetic Mastermind in Action.Appl Biochem Biotechnol. 2021 Jun;193(6):1701-1726. doi: 10.1007/s12010-021-03537-5. Epub 2021 Mar 10. Appl Biochem Biotechnol. 2021. PMID: 33694104 Review.
-
TET proteins and 5-methylcytosine oxidation in hematological cancers.Immunol Rev. 2015 Jan;263(1):6-21. doi: 10.1111/imr.12239. Immunol Rev. 2015. PMID: 25510268 Free PMC article. Review.
Cited by
-
Differential Expression of Steroid Hormone Receptors and Ten Eleven Translocation Proteins in Endometrial Cancer Cells.Front Oncol. 2022 Mar 11;12:763464. doi: 10.3389/fonc.2022.763464. eCollection 2022. Front Oncol. 2022. PMID: 35372016 Free PMC article.
-
Cancer Cell Phenotype Plasticity as a Driver of Immune Escape in Melanoma.Front Immunol. 2022 Mar 29;13:873116. doi: 10.3389/fimmu.2022.873116. eCollection 2022. Front Immunol. 2022. PMID: 35432344 Free PMC article. Review.
-
Epigenetic Regulators of DNA Cytosine Modification: Promising Targets for Cancer Therapy.Biomedicines. 2023 Feb 21;11(3):654. doi: 10.3390/biomedicines11030654. Biomedicines. 2023. PMID: 36979633 Free PMC article. Review.
-
Melanoma: Genetic Abnormalities, Tumor Progression, Clonal Evolution and Tumor Initiating Cells.Med Sci (Basel). 2017 Nov 20;5(4):28. doi: 10.3390/medsci5040028. Med Sci (Basel). 2017. PMID: 29156643 Free PMC article. Review.
-
Role of TET Dioxygenases and DNA Hydroxymethylation in Bisphenols-Stimulated Proliferation of Breast Cancer Cells.Environ Health Perspect. 2020 Feb;128(2):27008. doi: 10.1289/EHP5862. Epub 2020 Feb 27. Environ Health Perspect. 2020. PMID: 32105160 Free PMC article.
References
-
- Alonso SR, Tracey L, Ortiz P, Perez-Gomez B, Palacios J, Pollan M, Linares J, Serrano S, Saez-Castillo AI, Sanchez L, Pajares R, Sanchez-Aguilera A, Artiga MJ, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–60. doi: 10.1158/0008-5472.CAN-06-3481. - DOI - PubMed
-
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, Schadendorf D, Dummer R. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

