Time-scale of minor HIV-1 complex circulating recombinant forms from Central and West Africa
- PMID: 27852214
- PMCID: PMC5112642
- DOI: 10.1186/s12862-016-0824-8
Time-scale of minor HIV-1 complex circulating recombinant forms from Central and West Africa
Abstract
Background: Several HIV-1 circulating recombinant forms with a complex mosaic structure (CRFs_cpx) circulate in central and western African regions. Here we reconstruct the evolutionary history of some of these complex CRFs (09_cpx, 11_cpx, 13_cpx and 45_cpx) and further investigate the dissemination dynamic of the CRF11_cpx clade by using a Bayesian coalescent-based method.
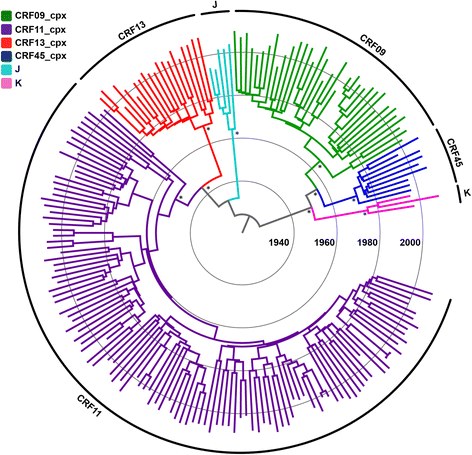
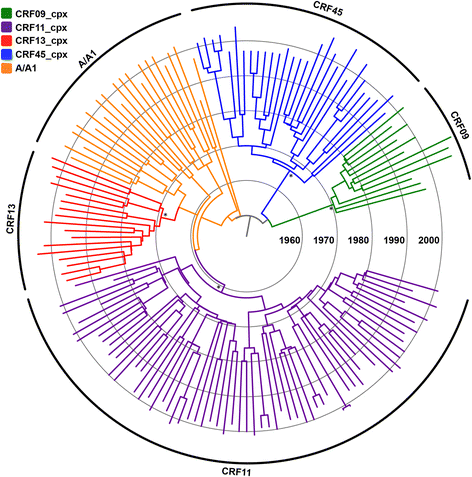
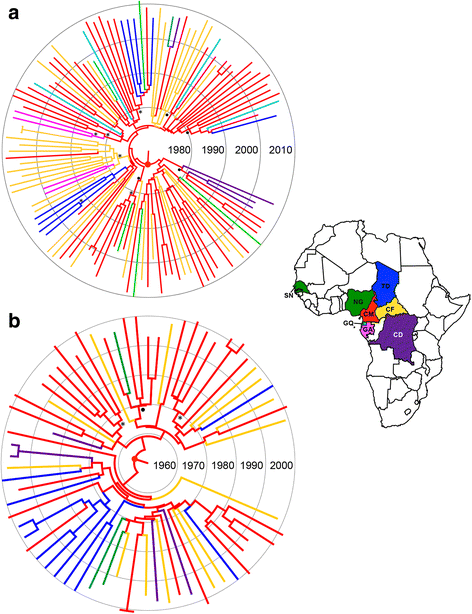
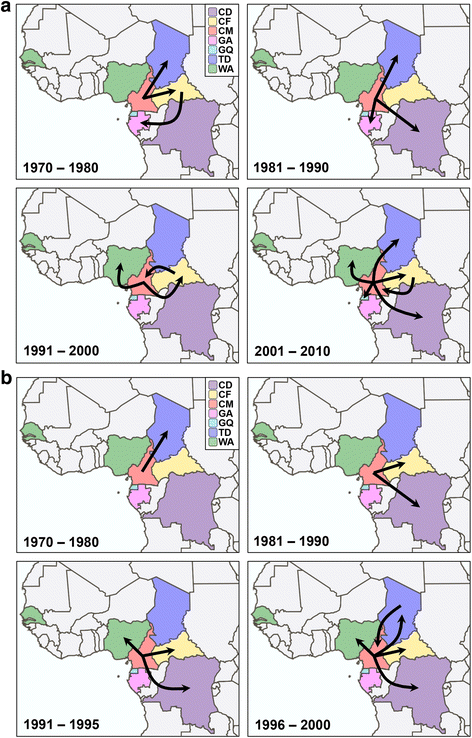
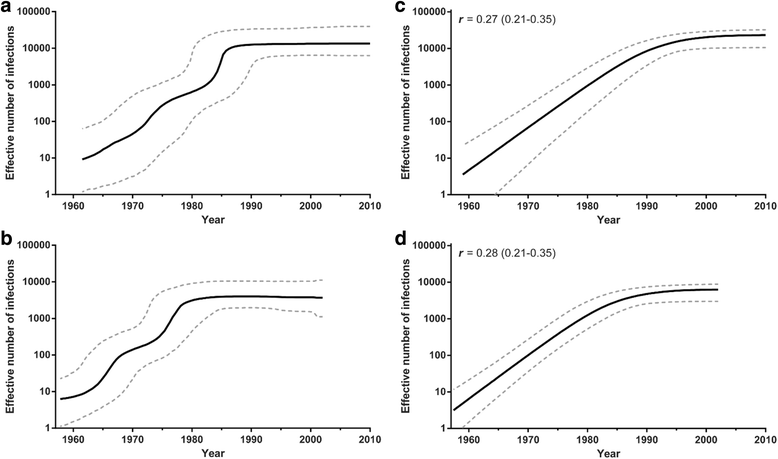
Results: The analysis of two HIV-1 datasets comprising 181 pol (36 CRF09_cpx, 116 CRF11_cpx, 20 CRF13_cpx and 9 CRF45_cpx) and 125 env (12 CRF09_cpx, 67 CRF11_cpx, 17 CRF13_cpx and 29 CRF45_cpx) sequences pointed to quite consistent onset dates for CRF09_cpx (~1966: 1958-1979), CRF11_cpx (~1957: 1950-1966) and CRF13_cpx (~1965: 1958-1973) clades; while some divergence was found for the estimated date of origin of CRF45_cpx clade [pol = 1970 (1964-1976); env = 1960 (1952-1969)]. Phylogeographic reconstructions indicate that the HIV-1 CRF11_cpx clade most probably emerged in Cameroon and from there it was first disseminated to the Central Africa Republic and Chad in the early 1970s and to other central and western African countries from the early 1980s onwards. Demographic reconstructions suggest that the CRF11_cpx epidemic grew between 1960 and 1990 with a median exponential growth rate of 0.27 year-1, and stabilized after.
Conclusions: These results reveal that HIV-1 CRFs_cpx clades have been circulating in Central Africa for a period comparable to other much more prevalent HIV-1 group M lineages. Cameroon was probably the epicenter of dissemination of the CRF11_cpx clade that seems to have experienced a long epidemic growth phase before stabilization. The epidemic growth of the CRF11_cpx clade was roughly comparable to other HIV-1 group M lineages circulating in Central Africa.
Keywords: Africa; Complex CRFs; HIV-1; Phylodynamics; Phylogeography.
Figures
Similar articles
-
Tracing the origin of a singular HIV-1 CRF45_cpx clade identified in Brazil.Infect Genet Evol. 2016 Dec;46:223-232. doi: 10.1016/j.meegid.2016.05.040. Epub 2016 May 31. Infect Genet Evol. 2016. PMID: 27259365
-
Spatiotemporal dynamics of the HIV-1 CRF06_cpx epidemic in Western Africa.AIDS. 2013 May 15;27(8):1313-20. doi: 10.1097/QAD.0b013e32835f1df4. AIDS. 2013. PMID: 23343915
-
Reconstructing the HIV-1 CRF02_AG and CRF06_cpx epidemics in Burkina Faso and West Africa using early samples.Infect Genet Evol. 2016 Dec;46:209-218. doi: 10.1016/j.meegid.2016.03.038. Epub 2016 Apr 6. Infect Genet Evol. 2016. PMID: 27063411
-
Prevalence and spatiotemporal dynamics of HIV-1 Circulating Recombinant Form 03_AB (CRF03_AB) in the Former Soviet Union countries.PLoS One. 2020 Oct 23;15(10):e0241269. doi: 10.1371/journal.pone.0241269. eCollection 2020. PLoS One. 2020. PMID: 33095842 Free PMC article.
-
Update on HIV-1 diversity in Africa: a decade in review.AIDS Rev. 2012 Apr-Jun;14(2):83-100. AIDS Rev. 2012. PMID: 22627605 Review.
Cited by
-
Genetic Diversity of HIV-1 in Krasnoyarsk Krai: Area with High Levels of HIV-1 Recombination in Russia.Biomed Res Int. 2020 Sep 10;2020:9057541. doi: 10.1155/2020/9057541. eCollection 2020. Biomed Res Int. 2020. PMID: 32964045 Free PMC article.
-
HIV-1 Unique Recombinant Forms Identified in Slovenia and Their Characterization by Near Full-Length Genome Sequencing.Viruses. 2020 Jan 3;12(1):63. doi: 10.3390/v12010063. Viruses. 2020. PMID: 31947872 Free PMC article.
-
The Role of Phylogenetics in Discerning HIV-1 Mixing among Vulnerable Populations and Geographic Regions in Sub-Saharan Africa: A Systematic Review.Viruses. 2021 Jun 19;13(6):1174. doi: 10.3390/v13061174. Viruses. 2021. PMID: 34205246 Free PMC article.
-
Geographically-stratified HIV-1 group M pol subtype and circulating recombinant form sequences.Sci Data. 2018 Jul 31;5:180148. doi: 10.1038/sdata.2018.148. Sci Data. 2018. PMID: 30063225 Free PMC article.
-
Elucidation of Early Evolution of HIV-1 Group M in the Congo Basin Using Computational Methods.Genes (Basel). 2021 Apr 2;12(4):517. doi: 10.3390/genes12040517. Genes (Basel). 2021. PMID: 33918115 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

