Genome-wide gene expression and RNA half-life measurements allow predictions of regulation and metabolic behavior in Methanosarcina acetivorans
- PMID: 27852217
- PMCID: PMC5112694
- DOI: 10.1186/s12864-016-3219-8
Genome-wide gene expression and RNA half-life measurements allow predictions of regulation and metabolic behavior in Methanosarcina acetivorans
Abstract
Background: While a few studies on the variations in mRNA expression and half-lives measured under different growth conditions have been used to predict patterns of regulation in bacterial organisms, the extent to which this information can also play a role in defining metabolic phenotypes has yet to be examined systematically. Here we present the first comprehensive study for a model methanogen.
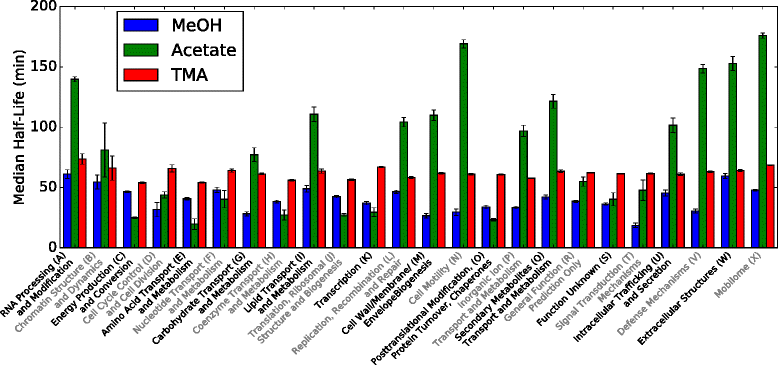
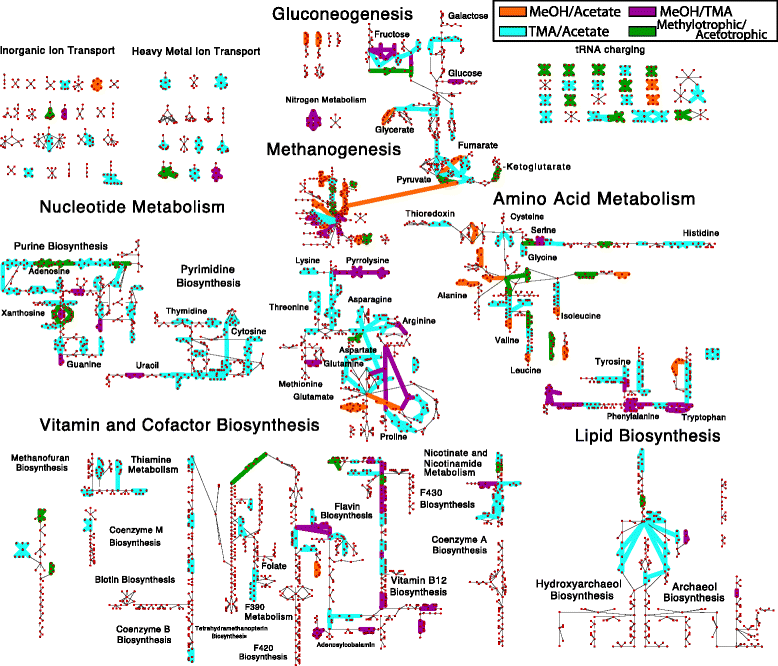
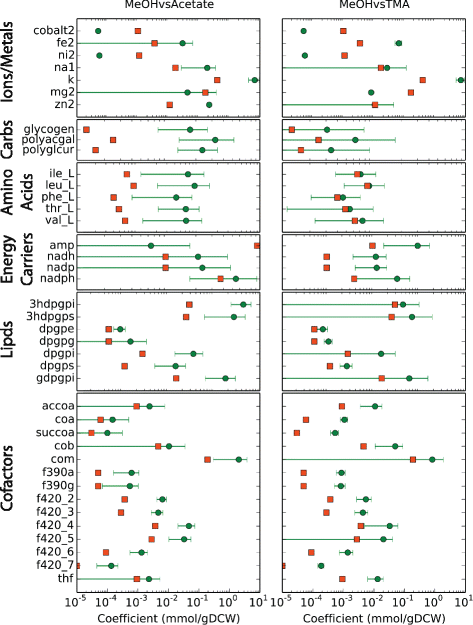
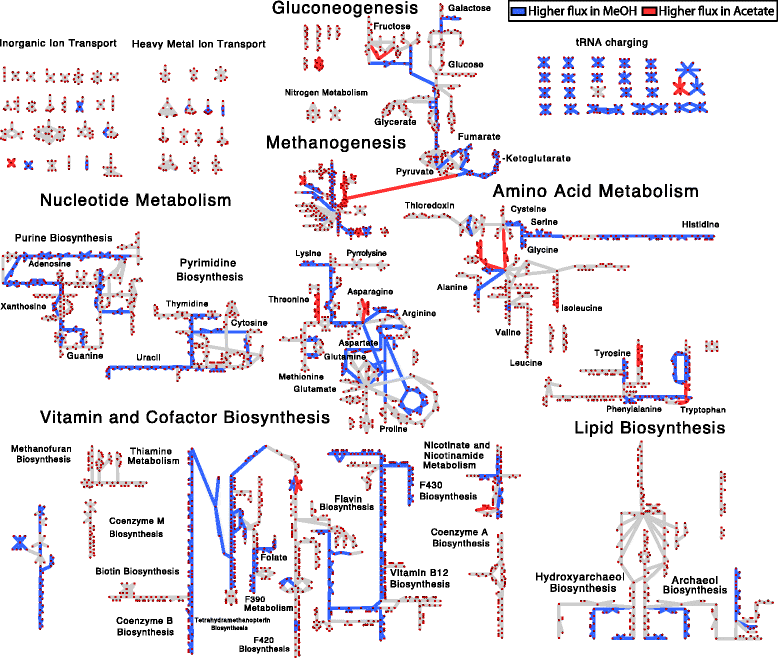
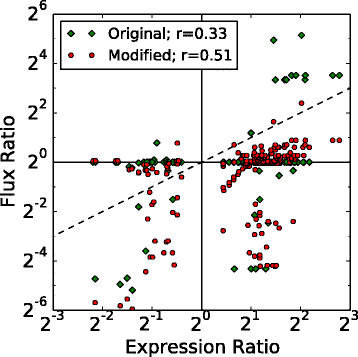
Results: We use expression and half-life data for the methanogen Methanosarcina acetivorans growing on fast- and slow-growth substrates to examine the regulation of its genes. Unlike Escherichia coli where only small shifts in half-lives were observed, we found that most mRNA have significantly longer half-lives for slow growth on acetate compared to fast growth on methanol or trimethylamine. Interestingly, half-life shifts are not uniform across functional classes of enzymes, suggesting the existence of a selective stabilization mechanism for mRNAs. Using the transcriptomics data we determined whether transcription or degradation rate controls the change in transcript abundance. Degradation was found to control abundance for about half of the metabolic genes underscoring its role in regulating metabolism. Genes involved in half of the metabolic reactions were found to be differentially expressed among the substrates suggesting the existence of drastically different metabolic phenotypes that extend beyond just the methanogenesis pathways. By integrating expression data with an updated metabolic model of the organism (iST807) significant differences in pathway flux and production of metabolites were predicted for the three growth substrates.
Conclusions: This study provides the first global picture of differential expression and half-lives for a class II methanogen, as well as provides the first evidence in a single organism that drastic genome-wide shifts in RNA half-lives can be modulated by growth substrate. We determined which genes in each metabolic pathway control the flux and classified them as regulated by transcription (e.g. transcription factor) or degradation (e.g. post-transcriptional modification). We found that more than half of genes in metabolism were controlled by degradation. Our results suggest that M. acetivorans employs extensive post-transcriptional regulation to optimize key metabolic steps, and more generally that degradation could play a much greater role in optimizing an organism's metabolism than previously thought.
Keywords: Degradational regulatory control; Differential pathway usage; Genome scale metabolic modeling; Metabolic phenotype; Methanogens; RNA half–lives.
Figures





Similar articles
-
Metabolic reconstruction of the archaeon methanogen Methanosarcina Acetivorans.BMC Syst Biol. 2011 Feb 15;5:28. doi: 10.1186/1752-0509-5-28. BMC Syst Biol. 2011. PMID: 21324125 Free PMC article.
-
Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon Methanosarcina acetivorans C2A.J Bacteriol. 2012 Feb;194(4):855-65. doi: 10.1128/JB.06040-11. Epub 2011 Dec 2. J Bacteriol. 2012. PMID: 22139506 Free PMC article.
-
Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the Archaean, Methanosarcina acetivorans strain C2A.BMC Microbiol. 2010 Feb 23;10:62. doi: 10.1186/1471-2180-10-62. BMC Microbiol. 2010. PMID: 20178638 Free PMC article.
-
A promoter-RBS library for fine-tuning gene expression in Methanosarcina acetivorans.Appl Environ Microbiol. 2024 Sep 18;90(9):e0109224. doi: 10.1128/aem.01092-24. Epub 2024 Aug 12. Appl Environ Microbiol. 2024. PMID: 39132998 Free PMC article.
-
Inhibiting transcription in cultured metazoan cells with actinomycin D to monitor mRNA turnover.Methods. 2019 Feb 15;155:77-87. doi: 10.1016/j.ymeth.2019.01.003. Epub 2019 Jan 6. Methods. 2019. PMID: 30625384 Free PMC article. Review.
Cited by
-
Transcriptional response of Methanosarcina acetivorans to repression of the energy-conserving methanophenazine: CoM-CoB heterodisulfide reductase enzyme HdrED.Microbiol Spectr. 2024 Oct 29;12(12):e0095724. doi: 10.1128/spectrum.00957-24. Online ahead of print. Microbiol Spectr. 2024. PMID: 39472004 Free PMC article.
-
In vivo structure probing of RNA in Archaea: novel insights into the ribosome structure of Methanosarcina acetivorans.RNA. 2023 Oct;29(10):1610-1620. doi: 10.1261/rna.079687.123. Epub 2023 Jul 25. RNA. 2023. PMID: 37491319 Free PMC article.
-
Short-Term Exposure of Paddy Soil Microbial Communities to Salt Stress Triggers Different Transcriptional Responses of Key Taxonomic Groups.Front Microbiol. 2017 Mar 28;8:400. doi: 10.3389/fmicb.2017.00400. eCollection 2017. Front Microbiol. 2017. PMID: 28400748 Free PMC article.
-
Hydrogenotrophic methanogenesis at 7-12 mbar by Methanosarcina barkeri under simulated martian atmospheric conditions.Sci Rep. 2025 Jan 22;15(1):2880. doi: 10.1038/s41598-025-86145-1. Sci Rep. 2025. PMID: 39843490 Free PMC article.
-
meCLICK-Seq, a Substrate-Hijacking and RNA Degradation Strategy for the Study of RNA Methylation.ACS Cent Sci. 2020 Dec 23;6(12):2196-2208. doi: 10.1021/acscentsci.0c01094. Epub 2020 Oct 29. ACS Cent Sci. 2020. PMID: 33376781 Free PMC article.
References
-
- Rogowska AT, Puchta O, Czarnecka AM, Kaniak A, Stepien PP, Golik P. Balance between Transcription and RNA Degradation Is Vital for Saccharomyces cerevisiae Mitochondria: Reduced Transcription Rescues the Phenotype of Deficient RNA Degradation. Mol Biol Cell. 2005;17(3):1184–93. doi: 10.1091/mbc.E05-08-0796. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

