RNA-seq based transcriptomic map reveals new insights into mouse salivary gland development and maturation
- PMID: 27852218
- PMCID: PMC5112738
- DOI: 10.1186/s12864-016-3228-7
RNA-seq based transcriptomic map reveals new insights into mouse salivary gland development and maturation
Abstract
Background: Mouse models have served a valuable role in deciphering various facets of Salivary Gland (SG) biology, from normal developmental programs to diseased states. To facilitate such studies, gene expression profiling maps have been generated for various stages of SG organogenesis. However these prior studies fall short of capturing the transcriptional complexity due to the limited scope of gene-centric microarray-based technology. Compared to microarray, RNA-sequencing (RNA-seq) offers unbiased detection of novel transcripts, broader dynamic range and high specificity and sensitivity for detection of genes, transcripts, and differential gene expression. Although RNA-seq data, particularly under the auspices of the ENCODE project, have covered a large number of biological specimens, studies on the SG have been lacking.
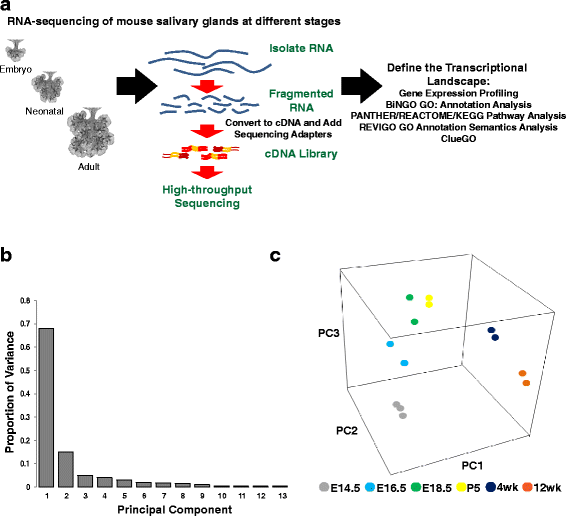
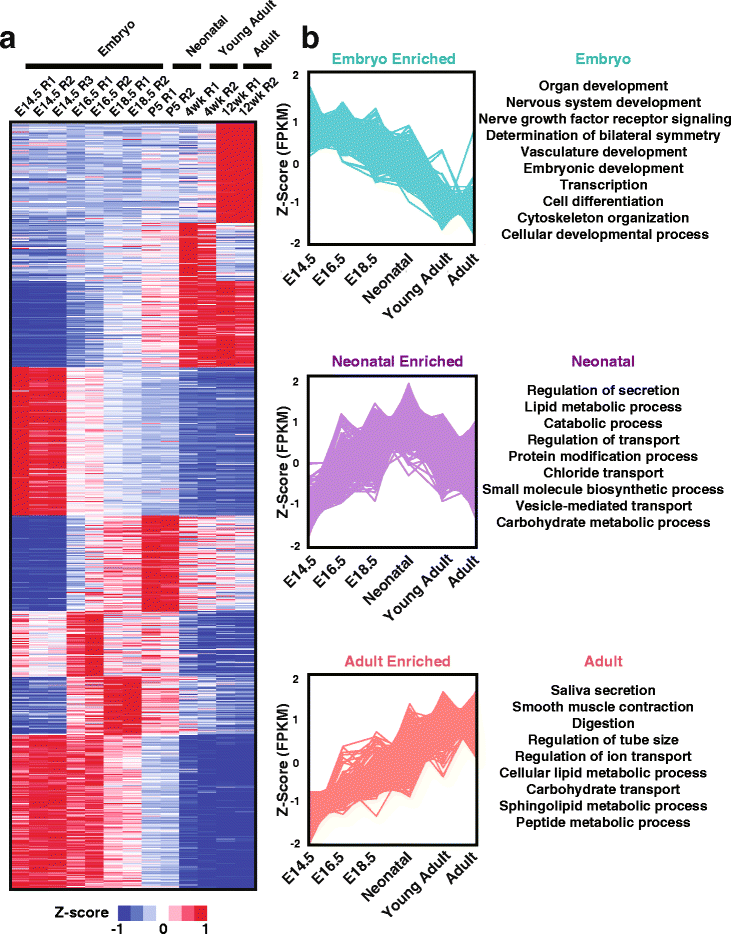
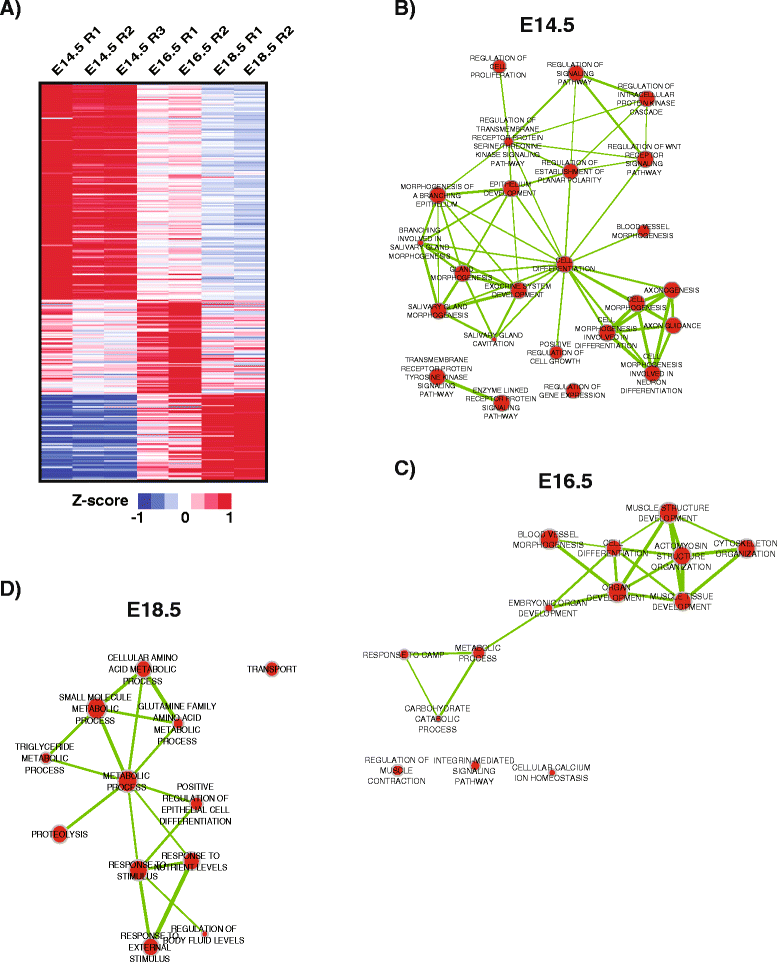
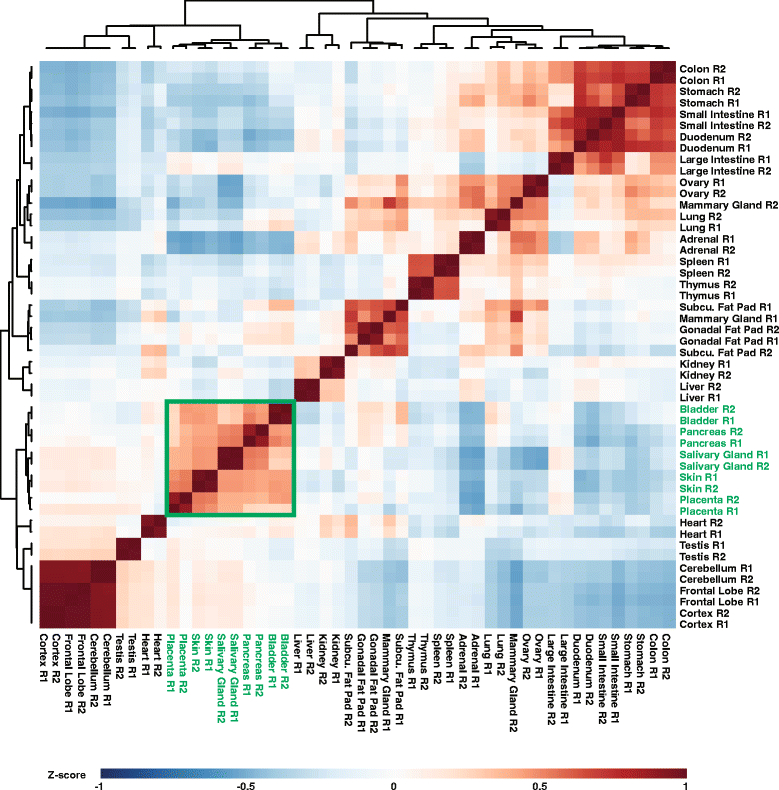
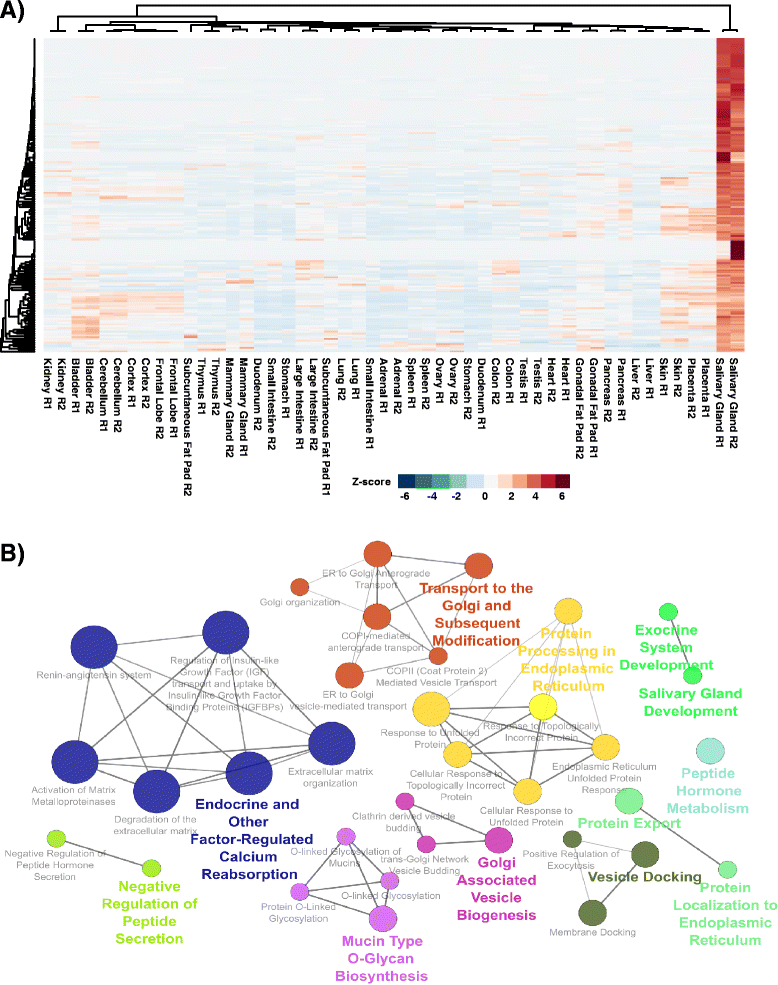
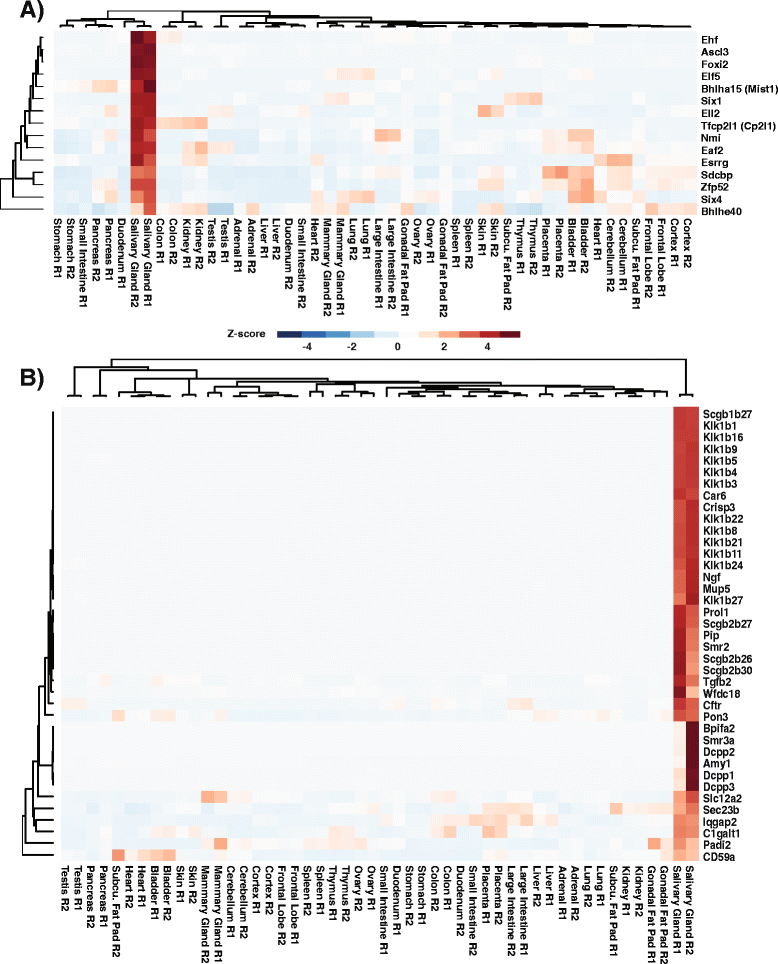
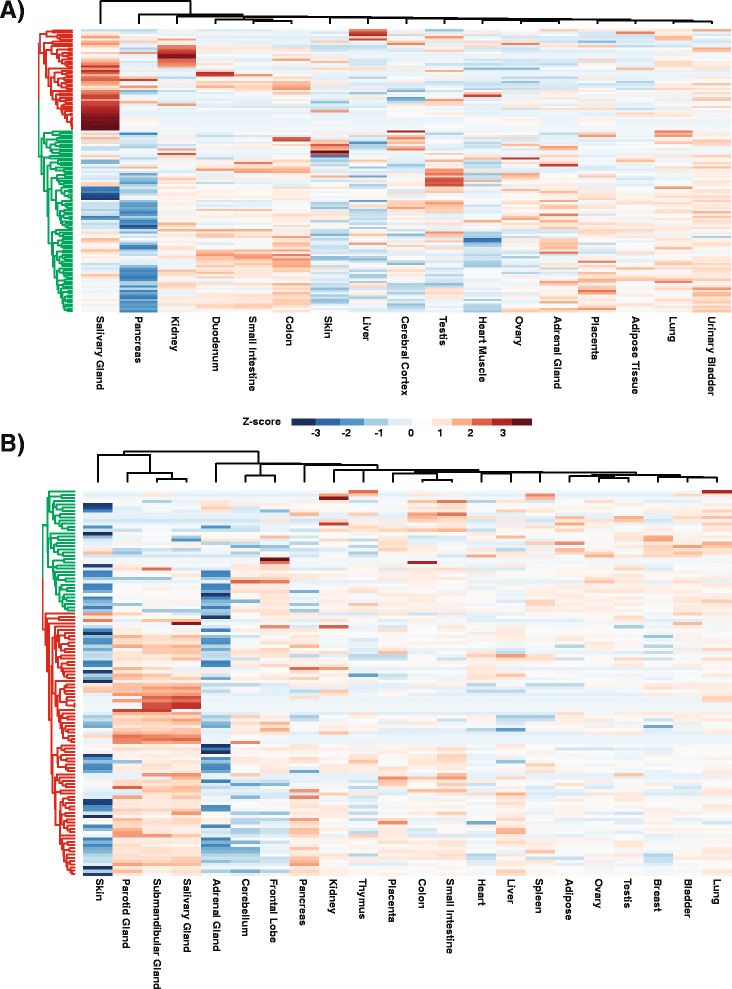
Results: To better appreciate the wide spectrum of gene expression profiles, we isolated RNA from mouse submandibular salivary glands at different embryonic and adult stages. In parallel, we processed RNA-seq data for 24 organs and tissues obtained from the mouse ENCODE consortium and calculated the average gene expression values. To identify molecular players and pathways likely to be relevant for SG biology, we performed functional gene enrichment analysis, network construction and hierarchal clustering of the RNA-seq datasets obtained from different stages of SG development and maturation, and other mouse organs and tissues. Our bioinformatics-based data analysis not only reaffirmed known modulators of SG morphogenesis but revealed novel transcription factors and signaling pathways unique to mouse SG biology and function. Finally we demonstrated that the unique SG gene signature obtained from our mouse studies is also well conserved and can demarcate features of the human SG transcriptome that is different from other tissues.
Conclusions: Our RNA-seq based Atlas has revealed a high-resolution cartographic view of the dynamic transcriptomic landscape of the mouse SG at various stages. These RNA-seq datasets will complement pre-existing microarray based datasets, including the Salivary Gland Molecular Anatomy Project by offering a broader systems-biology based perspective rather than the classical gene-centric view. Ultimately such resources will be valuable in providing a useful toolkit to better understand how the diverse cell population of the SG are organized and controlled during development and differentiation.
Keywords: Development; Gene signature; RNA-sequencing; Salivary gland.
Figures
References
-
- Nelson DA, Manhardt C, Kamath V, Sui Y, Santamaria-Pang A, Can A, Bello M, Corwin A, Dinn SR, Lazare M, et al. Quantitative single cell analysis of cell population dynamics during submandibular salivary gland development and differentiation. Biol Open. 2013;2(5):439–47. doi: 10.1242/bio.20134309. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

