Coevolution of paired receptors in Xenopus carcinoembryonic antigen-related cell adhesion molecule families suggests appropriation as pathogen receptors
- PMID: 27852220
- PMCID: PMC5112662
- DOI: 10.1186/s12864-016-3279-9
Coevolution of paired receptors in Xenopus carcinoembryonic antigen-related cell adhesion molecule families suggests appropriation as pathogen receptors
Abstract
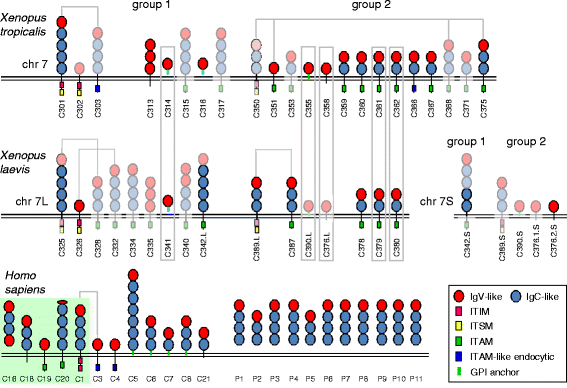
Background: In mammals, CEACAM1 and closely related members represent paired receptors with similar extracellular ligand-binding regions and cytoplasmic domains with opposing functions. Human CEACAM1 and CEACAM3 which have inhibitory ITIM/ITSM and activating ITAM-like motifs, respectively, in their cytoplasmic regions are such paired receptors. Various bacterial pathogens bind to CEACAM1 on epithelial and immune cells facilitating both entry into the host and down-regulation of the immune response whereas interaction with granulocyte-specific CEACAM3 leads to their uptake and destruction. It is unclear whether paired CEACAM receptors also exist in other vertebrate clades.
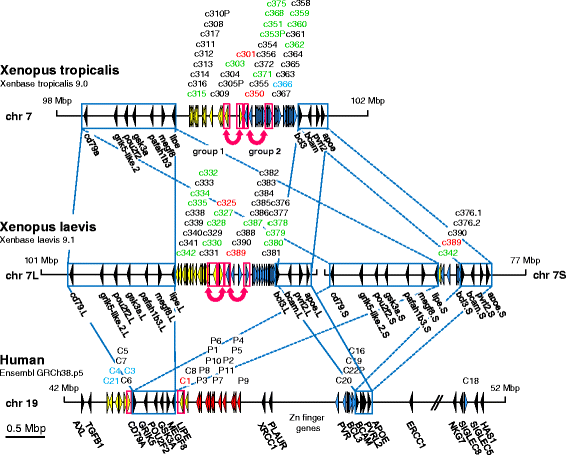
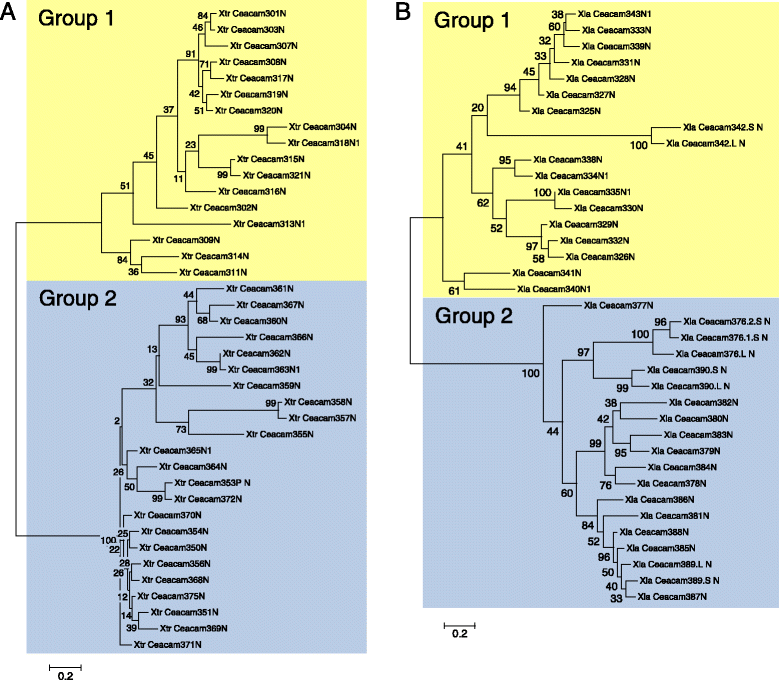
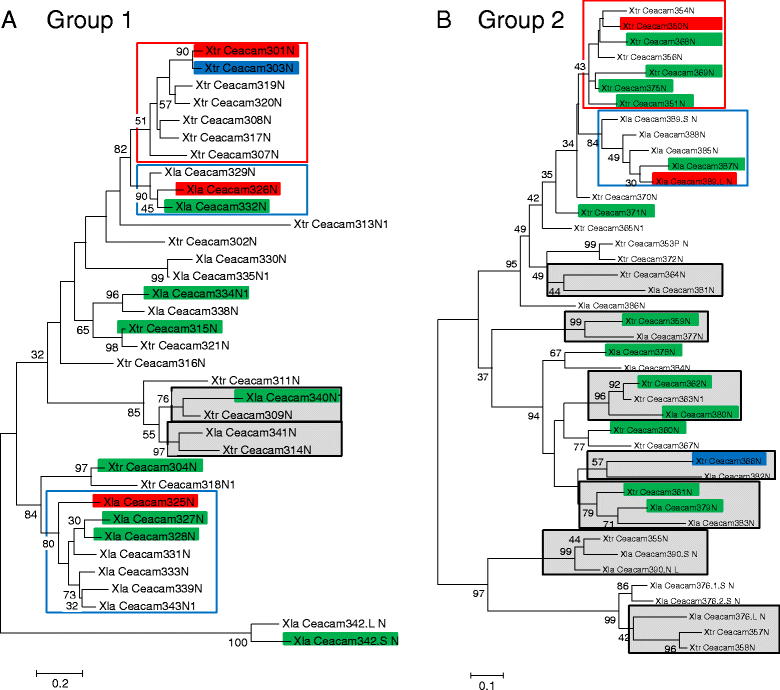
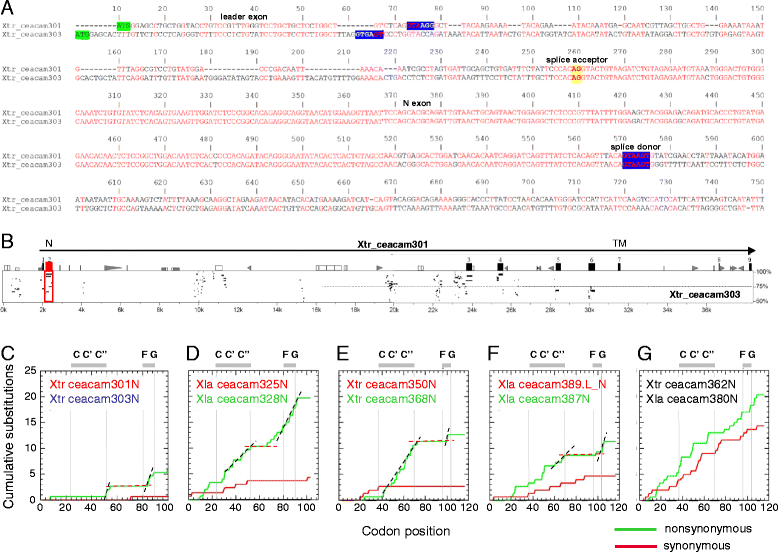
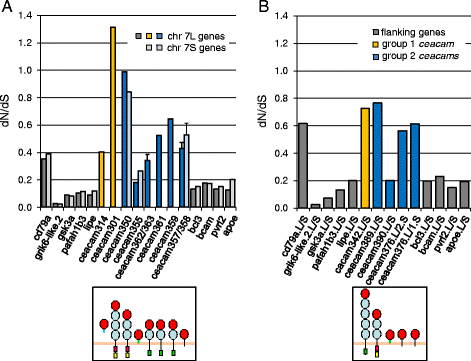
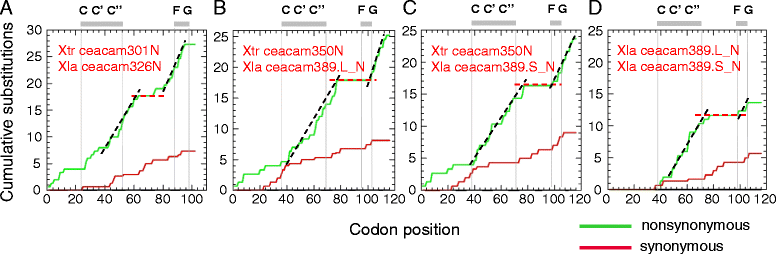
Results: We identified more than 80 ceacam genes in Xenopus tropicalis and X. laevis. They consist of two subgroups containing one or two putative paired receptor pairs each. Analysis of genomic sequences of paired receptors provide evidence that their highly similar ligand binding domains were adjusted by recent gene conversion events. In contrast, selection for diversification is observed among inhibitory receptor orthologs of the two frogs which split some 60 million years ago. The allotetraploid X. laevis arose later by hybridization of two closely related species. Interestingly, despite the conservation of the genomic landscape surrounding the homeologous ceacam loci only one locus resembles the one found in X. tropicalis. From the second X. laevis locus more than 80 % of the ceacam genes were lost including 5 of the 6 paired receptor genes. This suggests that once the gene for one of the paired receptors is lost the remaining gene cluster degrades rapidly probably due to lack of selection pressure exerted by pathogens.
Conclusions: The presence of paired receptors and selection for diversification suggests that also in amphibians CEACAM1-related inhibitory proteins are or were used as pathogen receptors.
Keywords: Allotetraploid; Amphibia; Carcinoembryonic antigen-related cell-cell adhesion molecule (CEACAM); ITAM; ITIM; Immunoglobulin superfamily; Paired receptors; Positive selection; Xenopus.
Figures
Similar articles
-
Evolution of CEACAM pathogen decoy receptors in primates.Eur J Clin Invest. 2024 Dec;54 Suppl 2(Suppl 2):e14356. doi: 10.1111/eci.14356. Eur J Clin Invest. 2024. PMID: 39674876 Free PMC article.
-
Coevolution of activating and inhibitory receptors within mammalian carcinoembryonic antigen families.BMC Biol. 2010 Feb 4;8:12. doi: 10.1186/1741-7007-8-12. BMC Biol. 2010. PMID: 20132533 Free PMC article.
-
Species-specific evolution of immune receptor tyrosine based activation motif-containing CEACAM1-related immune receptors in the dog.BMC Evol Biol. 2007 Oct 18;7:196. doi: 10.1186/1471-2148-7-196. BMC Evol Biol. 2007. PMID: 17945019 Free PMC article.
-
HemITAM signaling by CEACAM3, a human granulocyte receptor recognizing bacterial pathogens.Arch Biochem Biophys. 2012 Aug 1;524(1):77-83. doi: 10.1016/j.abb.2012.03.020. Epub 2012 Mar 24. Arch Biochem Biophys. 2012. PMID: 22469950 Review.
-
CEACAM3: an innate immune receptor directed against human-restricted bacterial pathogens.Int J Med Microbiol. 2008 Oct;298(7-8):553-60. doi: 10.1016/j.ijmm.2008.04.005. Epub 2008 Jul 7. Int J Med Microbiol. 2008. PMID: 18606569 Review.
Cited by
-
Alternative splicing after gene duplication drives CEACAM1-paralog diversification in the horse.BMC Evol Biol. 2018 Mar 15;18(1):32. doi: 10.1186/s12862-018-1145-x. BMC Evol Biol. 2018. PMID: 29544443 Free PMC article.
-
Evolution of CEACAM pathogen decoy receptors in primates.Eur J Clin Invest. 2024 Dec;54 Suppl 2(Suppl 2):e14356. doi: 10.1111/eci.14356. Eur J Clin Invest. 2024. PMID: 39674876 Free PMC article.
-
Transcriptome of the Southern Muriqui Brachyteles arachnoides (Primates:Platyrrhini), a Critically Endangered New World Monkey: Evidence of Adaptive Evolution.Front Genet. 2020 Jul 31;11:831. doi: 10.3389/fgene.2020.00831. eCollection 2020. Front Genet. 2020. PMID: 32849820 Free PMC article.
-
Structural characterization of a dimeric complex between the short cytoplasmic domain of CEACAM1 and the pseudo tetramer of S100A10-Annexin A2 using NMR and molecular dynamics.Biochim Biophys Acta Biomembr. 2021 Jan 1;1863(1):183451. doi: 10.1016/j.bbamem.2020.183451. Epub 2020 Aug 21. Biochim Biophys Acta Biomembr. 2021. PMID: 32835655 Free PMC article.
-
Natural selection supports escape from concerted evolution of a recently duplicated CEACAM1 paralog in the ruminant CEA gene family.Sci Rep. 2020 Feb 25;10(1):3404. doi: 10.1038/s41598-020-60425-4. Sci Rep. 2020. PMID: 32099040 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

