Differential gene expression in Varroa jacobsoni mites following a host shift to European honey bees (Apis mellifera)
- PMID: 27852222
- PMCID: PMC5112721
- DOI: 10.1186/s12864-016-3130-3
Differential gene expression in Varroa jacobsoni mites following a host shift to European honey bees (Apis mellifera)
Abstract
Background: Varroa mites are widely considered the biggest honey bee health problem worldwide. Until recently, Varroa jacobsoni has been found to live and reproduce only in Asian honey bee (Apis cerana) colonies, while V. destructor successfully reproduces in both A. cerana and A. mellifera colonies. However, we have identified an island population of V. jacobsoni that is highly destructive to A. mellifera, the primary species used for pollination and honey production. The ability of these populations of mites to cross the host species boundary potentially represents an enormous threat to apiculture, and is presumably due to genetic variation that exists among populations of V. jacobsoni that influences gene expression and reproductive status. In this work, we investigate differences in gene expression between populations of V. jacobsoni reproducing on A. cerana and those either reproducing or not capable of reproducing on A. mellifera, in order to gain insight into differences that allow V. jacobsoni to overcome its normal species tropism.
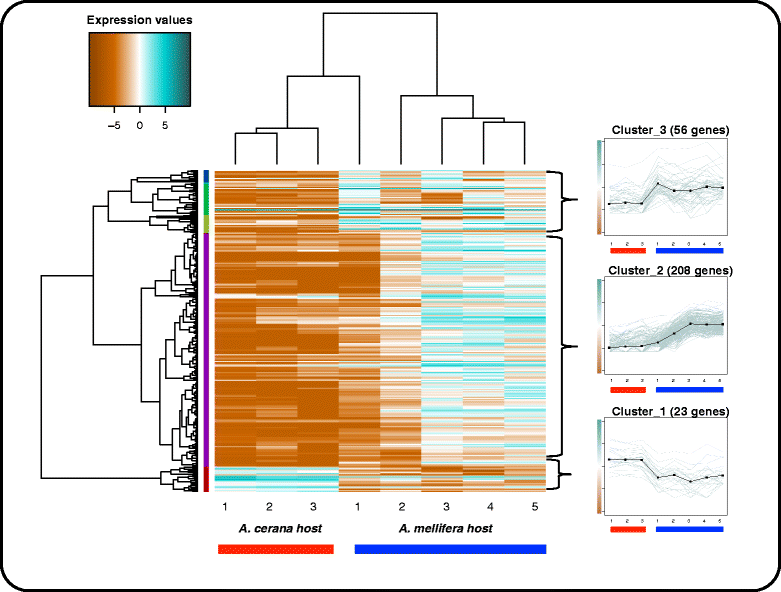
Results: We sequenced and assembled a de novo transcriptome of V. jacobsoni. We also performed a differential gene expression analysis contrasting biological replicates of V. jacobsoni populations that differ in their ability to reproduce on A. mellifera. Using the edgeR, EBSeq and DESeq R packages for differential gene expression analysis, we found 287 differentially expressed genes (FDR ≤ 0.05), of which 91% were up regulated in mites reproducing on A. mellifera. In addition, mites found reproducing on A. mellifera showed substantially more variation in expression among replicates. We searched for orthologous genes in public databases and were able to associate 100 of these 287 differentially expressed genes with a functional description.
Conclusions: There is differential gene expression between the two mite groups, with more variation in gene expression among mites that were able to reproduce on A. mellifera. A small set of genes showed reduced expression in mites on the A. mellifera host, including putative transcription factors and digestive tract developmental genes. The vast majority of differentially expressed genes were up-regulated in this host. This gene set showed enrichment for genes associated with mitochondrial respiratory function and apoptosis, suggesting that mites on this host may be experiencing higher stress, and may be less optimally adapted to parasitize it. Some genes involved in reproduction and oogenesis were also overexpressed, which should be further studied in regards to this host shift.
Keywords: Apis cerana; Apis mellifera; Asian honey bee; European honey bee; RNA-Seq; Transcriptome; Varroa destructor; Varroa jacobsoni.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

