Genomic differentiation among wild cyanophages despite widespread horizontal gene transfer
- PMID: 27852226
- PMCID: PMC5112629
- DOI: 10.1186/s12864-016-3286-x
Genomic differentiation among wild cyanophages despite widespread horizontal gene transfer
Abstract
Background: Genetic recombination is a driving force in genome evolution. Among viruses it has a dual role. For genomes with higher fitness, it maintains genome integrity in the face of high mutation rates. Conversely, for genomes with lower fitness, it provides immediate access to sequence space that cannot be reached by mutation alone. Understanding how recombination impacts the cohesion and dissolution of individual whole genomes within viral sequence space is poorly understood across double-stranded DNA bacteriophages (a.k.a phages) due to the challenges of obtaining appropriately scaled genomic datasets.
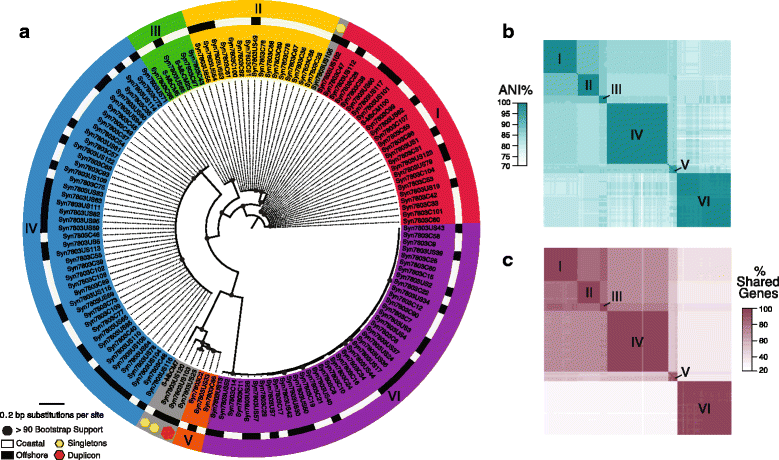
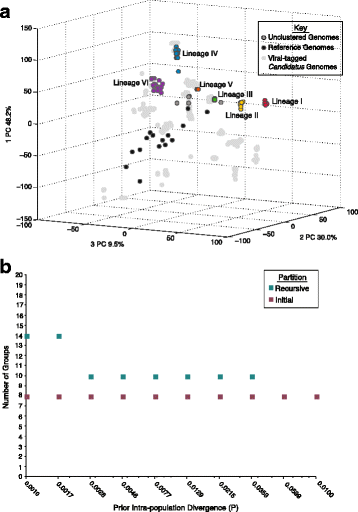
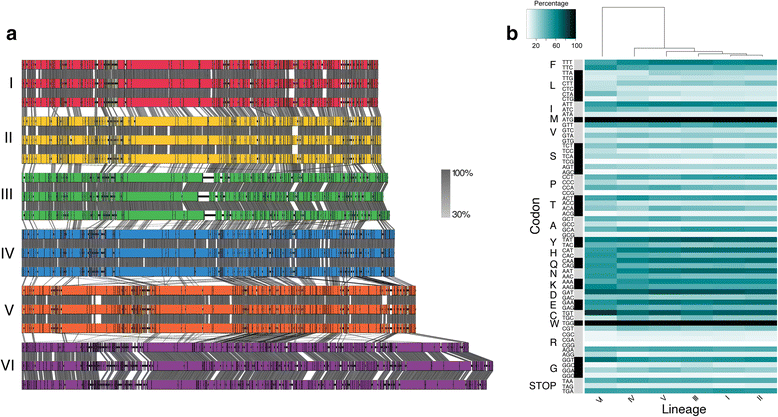
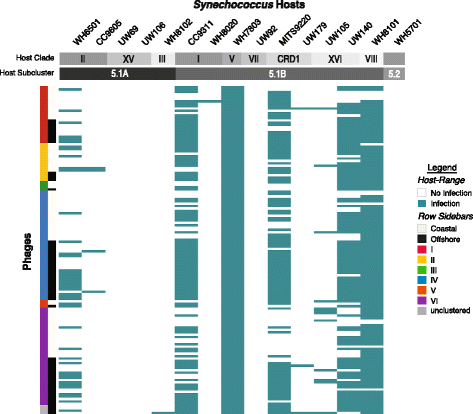
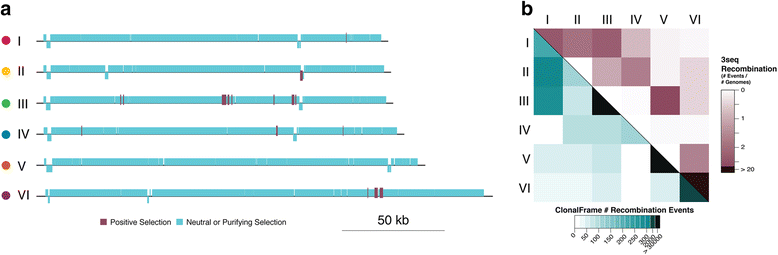
Results: Here we explore the role of recombination in both maintaining and differentiating whole genomes of 142 wild double-stranded DNA marine cyanophages. Phylogenomic analysis across the 51 core genes revealed ten lineages, six of which were well represented. These phylogenomic lineages represent discrete genotypic populations based on comparisons of intra- and inter- lineage shared gene content, genome-wide average nucleotide identity, as well as detected gaps in the distribution of pairwise differences between genomes. McDonald-Kreitman selection tests identified putative niche-differentiating genes under positive selection that differed across the six well-represented genotypic populations and that may have driven initial divergence. Concurrent with patterns of recombination of discrete populations, recombination analyses of both genic and intergenic regions largely revealed decreased genetic exchange across individual genomes between relative to within populations.
Conclusions: These findings suggest that discrete double-stranded DNA marine cyanophage populations occur in nature and are maintained by patterns of recombination akin to those observed in bacteria, archaea and in sexual eukaryotes.
Keywords: Bacteriophage; Cyanophage; Double-stranded DNA; Evolution; Phage; Species; Virus.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

