MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines
- PMID: 27852821
- PMCID: PMC5217702
- DOI: 10.1074/jbc.M116.750950
MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines
Abstract
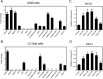
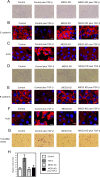
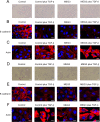
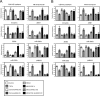
Histone methylation is implicated in a number of biological and pathological processes, including cancer development. In this study, we investigated the molecular mechanism for the recruitment of Polycomb repressive complex-2 (PRC2) and its accessory component, JARID2, to chromatin, which regulates methylation of lysine 27 of histone H3 (H3K27), during epithelial-mesenchymal transition (EMT) of cancer cells. The expression of MEG3 long noncoding RNA (lncRNA), which could interact with JARID2, was clearly increased during transforming growth factor-β (TGF-β)-induced EMT of human lung cancer cell lines. Knockdown of MEG3 inhibited TGF-β-mediated changes in cell morphology and cell motility characteristic of EMT and counteracted TGF-β-dependent changes in the expression of EMT-related genes such as CDH1, ZEB family, and the microRNA-200 family. Overexpression of MEG3 influenced the expression of these genes and enhanced the effects of TGF-β in their expressions. Chromatin immunoprecipitation (ChIP) revealed that MEG3 regulated the recruitment of JARID2 and EZH2 and histone H3 methylation on the regulatory regions of CDH1 and microRNA-200 family genes for transcriptional repression. RNA immunoprecipitation and chromatin isolation by RNA purification assays indicated that MEG3 could associate with JARID2 and the regulatory regions of target genes to recruit the complex. This study demonstrated a crucial role of MEG3 lncRNA in the epigenetic regulation of the EMT process in lung cancer cells.
Keywords: cancer biology; epithelial-mesenchymal transition (EMT); histone methylation; long noncoding RNA (long ncRNA, lncRNA); transcription regulation.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






Similar articles
-
MEG8 long noncoding RNA contributes to epigenetic progression of the epithelial-mesenchymal transition of lung and pancreatic cancer cells.J Biol Chem. 2018 Nov 23;293(47):18016-18030. doi: 10.1074/jbc.RA118.004006. Epub 2018 Sep 27. J Biol Chem. 2018. PMID: 30262664 Free PMC article.
-
JARID2 is involved in transforming growth factor-beta-induced epithelial-mesenchymal transition of lung and colon cancer cell lines.PLoS One. 2014 Dec 26;9(12):e115684. doi: 10.1371/journal.pone.0115684. eCollection 2014. PLoS One. 2014. PMID: 25542019 Free PMC article.
-
KDM2B is involved in the epigenetic regulation of TGF-β-induced epithelial-mesenchymal transition in lung and pancreatic cancer cell lines.J Biol Chem. 2021 Jan-Jun;296:100213. doi: 10.1074/jbc.RA120.015502. Epub 2020 Dec 24. J Biol Chem. 2021. PMID: 33779563 Free PMC article.
-
Epigenetic regulation in human melanoma: past and future.Epigenetics. 2015;10(2):103-21. doi: 10.1080/15592294.2014.1003746. Epigenetics. 2015. PMID: 25587943 Free PMC article. Review.
-
RNA regulatory mechanisms controlling TGF-β signaling and EMT in cancer.Semin Cancer Biol. 2024 Jul;102-103:4-16. doi: 10.1016/j.semcancer.2024.06.001. Epub 2024 Jun 23. Semin Cancer Biol. 2024. PMID: 38917876 Review.
Cited by
-
Long Non-Coding RNA in the Pathogenesis of Cancers.Cells. 2019 Sep 1;8(9):1015. doi: 10.3390/cells8091015. Cells. 2019. PMID: 31480503 Free PMC article. Review.
-
MEG3 is increased in idiopathic pulmonary fibrosis and regulates epithelial cell differentiation.JCI Insight. 2018 Sep 6;3(17):e122490. doi: 10.1172/jci.insight.122490. eCollection 2018 Sep 6. JCI Insight. 2018. PMID: 30185671 Free PMC article.
-
The Biological Roles and Molecular Mechanisms of Long Non-Coding RNA MEG3 in the Hallmarks of Cancer.Cancers (Basel). 2022 Dec 7;14(24):6032. doi: 10.3390/cancers14246032. Cancers (Basel). 2022. PMID: 36551518 Free PMC article. Review.
-
TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis.Int J Mol Sci. 2019 Jun 5;20(11):2767. doi: 10.3390/ijms20112767. Int J Mol Sci. 2019. PMID: 31195692 Free PMC article. Review.
-
LncRNAs: Proverbial Genomic "Junk" or Key Epigenetic Regulators During Cardiac Fibrosis in Diabetes?Front Cardiovasc Med. 2018 Apr 4;5:28. doi: 10.3389/fcvm.2018.00028. eCollection 2018. Front Cardiovasc Med. 2018. PMID: 29670886 Free PMC article.
References
-
- Kooistra S. M., and Helin K. (2012) Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13, 297–311 - PubMed
-
- Suzuki T., Shen H., Akagi K., Morse H. C., Malley J. D., Naiman D. Q., Jenkins N. A., and Copeland N. G. (2002) New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32, 166–174 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

