Mapping of Functional Subdomains in the Terminal Protein Domain of Hepatitis B Virus Polymerase
- PMID: 27852858
- PMCID: PMC5244320
- DOI: 10.1128/JVI.01785-16
Mapping of Functional Subdomains in the Terminal Protein Domain of Hepatitis B Virus Polymerase
Abstract
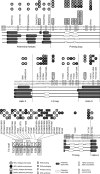
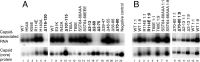
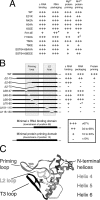
Hepatitis B virus (HBV) encodes a multifunction reverse transcriptase or polymerase (P), which is composed of several domains. The terminal protein (TP) domain is unique to HBV and related hepadnaviruses and is required for specifically binding to the viral pregenomic RNA (pgRNA). Subsequently, the TP domain is necessary for pgRNA packaging into viral nucleocapsids and the initiation of viral reverse transcription for conversion of the pgRNA to viral DNA. Uniquely, the HBV P protein initiates reverse transcription via a protein priming mechanism using the TP domain as a primer. No structural homologs or high-resolution structure exists for the TP domain. Secondary structure prediction identified three disordered loops in TP with highly conserved sequences. A meta-analysis of mutagenesis studies indicated these predicted loops are almost exclusively where functionally important residues are located. Newly constructed TP mutations revealed a priming loop in TP which plays a specific role in protein-primed DNA synthesis beyond simply harboring the site of priming. Substitutions of potential sites of phosphorylation surrounding the priming site demonstrated that these residues are involved in interactions critical for priming but are unlikely to be phosphorylated during viral replication. Furthermore, the first 13 and 66 TP residues were shown to be dispensable for protein priming and pgRNA binding, respectively. Combining current and previous mutagenesis work with sequence analysis has increased our understanding of TP structure and functions by mapping specific functions to distinct predicted secondary structures and will facilitate antiviral targeting of this unique domain.
Importance: HBV is a major cause of viral hepatitis, liver cirrhosis, and hepatocellular carcinoma. One important feature of this virus is its polymerase, the enzyme used to create the DNA genome from a specific viral RNA by reverse transcription. One region of this polymerase, the TP domain, is required for association with the viral RNA and production of the DNA genome. Targeting the TP domain for antiviral development is difficult due to the lack of homology to other proteins and high-resolution structure. This study mapped the TP functions according to predicted secondary structure, where it folds into alpha helices or unstructured loops. Three predicted loops were found to be the most important regions functionally and the most conserved evolutionarily. Identification of these functional subdomains in TP will facilitate its targeting for antiviral development.
Keywords: DNA polymerase; RNA binding; hepadnavirus; hepatitis B virus; protein priming; reverse transcriptase; terminal protein.
Copyright © 2017 American Society for Microbiology.
Figures

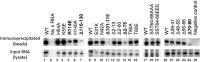

Similar articles
-
RNA-Binding Motif Protein 24 (RBM24) Is Involved in Pregenomic RNA Packaging by Mediating Interaction between Hepatitis B Virus Polymerase and the Epsilon Element.J Virol. 2019 Mar 5;93(6):e02161-18. doi: 10.1128/JVI.02161-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626666 Free PMC article.
-
Molecular, Evolutionary, and Structural Analysis of the Terminal Protein Domain of Hepatitis B Virus Polymerase, a Potential Drug Target.Viruses. 2020 May 22;12(5):570. doi: 10.3390/v12050570. Viruses. 2020. PMID: 32455999 Free PMC article. Review.
-
Comparative analysis of hepatitis B virus polymerase sequences required for viral RNA binding, RNA packaging, and protein priming.J Virol. 2014 Feb;88(3):1564-72. doi: 10.1128/JVI.02852-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227865 Free PMC article.
-
A conserved arginine residue in the terminal protein domain of hepatitis B virus polymerase is critical for RNA pre-genome encapsidation.J Gen Virol. 2011 Aug;92(Pt 8):1809-1816. doi: 10.1099/vir.0.031914-0. Epub 2011 Apr 27. J Gen Virol. 2011. PMID: 21525211
-
Hepatitis B viruses: reverse transcription a different way.Virus Res. 2008 Jun;134(1-2):235-49. doi: 10.1016/j.virusres.2007.12.024. Epub 2008 Mar 12. Virus Res. 2008. PMID: 18339439 Review.
Cited by
-
Novel Genetic Variants of Hepatitis B Virus in Fulminant Hepatitis.J Pathog. 2017;2017:1231204. doi: 10.1155/2017/1231204. Epub 2017 Dec 19. J Pathog. 2017. PMID: 29410920 Free PMC article.
-
Higher Rates of Viral Evolution in Chronic Hepatitis B Patients Linked to Predicted T Cell Epitopes.Viruses. 2025 May 8;17(5):684. doi: 10.3390/v17050684. Viruses. 2025. PMID: 40431695 Free PMC article.
-
Molecular Characterization of Hepatitis B Virus in People Living with HIV in Rural and Peri-Urban Communities in Botswana.Biomedicines. 2024 Jul 14;12(7):1561. doi: 10.3390/biomedicines12071561. Biomedicines. 2024. PMID: 39062134 Free PMC article.
-
Phylogenetic evidence supporting the nonenveloped nature of hepadnavirus ancestors.Proc Natl Acad Sci U S A. 2024 Nov 5;121(45):e2415631121. doi: 10.1073/pnas.2415631121. Epub 2024 Oct 29. Proc Natl Acad Sci U S A. 2024. PMID: 39471221 Free PMC article.
-
Protein phosphatase 1 catalyzes HBV core protein dephosphorylation and is co-packaged with viral pregenomic RNA into nucleocapsids.PLoS Pathog. 2020 Jul 23;16(7):e1008669. doi: 10.1371/journal.ppat.1008669. eCollection 2020 Jul. PLoS Pathog. 2020. PMID: 32702076 Free PMC article.
References
-
- Hu J. 2016. Hepatitis B virus virology and replication, p 1–34. In Liaw Y-F, Zoulim F (ed), Hepatitis B virus in human diseases. Humana Press, Springer, New York, New York.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

