Unmasking the U2AF homology motif family: a bona fide protein-protein interaction motif in disguise
- PMID: 27852923
- PMCID: PMC5113200
- DOI: 10.1261/rna.057950.116
Unmasking the U2AF homology motif family: a bona fide protein-protein interaction motif in disguise
Abstract
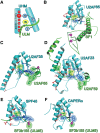
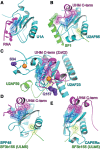
U2AF homology motifs (UHM) that recognize U2AF ligand motifs (ULM) are an emerging family of protein-protein interaction modules. UHM-ULM interactions recur in pre-mRNA splicing factors including U2AF1 and SF3b1, which are frequently mutated in myelodysplastic syndromes. The core topology of the UHM resembles an RNA recognition motif and is often mistakenly classified within this large family. Here, we unmask the charade and review recent discoveries of UHM-ULM modules for protein-protein interactions. Diverse polypeptide extensions and selective phosphorylation of UHM and ULM family members offer new molecular mechanisms for the assembly of specific partners in the early-stage spliceosome.
Keywords: RRM; SF3B1; U2AF1; UHM; protein–protein interaction.
© 2016 Loerch and Kielkopf; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

References
-
- Avis JM, Allain FH, Howe PW, Varani G, Nagai K, Neuhaus D. 1996. Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J Mol Biol 257: 398–411. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
