Solution structure of the 5'-terminal hairpin of the 7SK small nuclear RNA
- PMID: 27852926
- PMCID: PMC5113205
- DOI: 10.1261/rna.056523.116
Solution structure of the 5'-terminal hairpin of the 7SK small nuclear RNA
Abstract
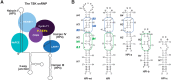
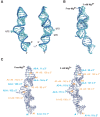
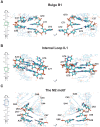
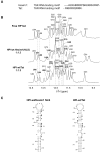
The small nuclear 7SK RNA regulates RNA polymerase II (RNA Pol II) transcription, by sequestering and inhibiting the positive transcription elongation factor b (P-TEFb). P-TEFb is stored in the 7SK ribonucleoprotein (RNP) that contains the three nuclear proteins Hexim1, LaRP7, and MePCE. P-TEFb interacts with the protein Hexim1 and the 7SK RNA. Once P-TEFb is released from the 7SK RNP, it activates transcription by phosphorylating the C-terminal domain of RNA Pol II. P-TEFb also plays a crucial role in the replication of the human immunodeficiency virus HIV-1, through its recruitment by the viral transactivator Tat. Previous work demonstrated that the protein Tat promotes the release of P-TEFb from the 7SK RNP through direct binding to the 7SK RNA. Hexim1 and Tat proteins both comprise conserved and similar arginine-rich motifs that were identified to bind the 7SK RNA at a repeated GAUC site located at the top of the 5'-terminal hairpin (HPI). Here, we report the solution structure of this region as determined by nuclear magnetic resonance, to identify HPI structural features recognized by Hexim1 and Tat. The HPI solution structure displays an elongated shape featuring four helical segments interrupted by one internal loop and three bulges with distinct folds. In particular, the repeated GAUC motif adopts a pre-organized geometry. Our results suggest that the binding of Hexim1 and Tat to the 7SK RNA could originate from a conformational selection of this motif, highlighting how RNA local structure could lead to an adaptive recognition of their partners.
Keywords: 7SK; Hexim; NMR; RNA; TAR; Tat.
© 2016 Bourbigot et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



Similar articles
-
A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb.Mol Cell Biol. 2004 Jun;24(12):5094-105. doi: 10.1128/MCB.24.12.5094-5105.2004. Mol Cell Biol. 2004. PMID: 15169877 Free PMC article.
-
Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat.PLoS Pathog. 2010 Oct 14;6(10):e1001152. doi: 10.1371/journal.ppat.1001152. PLoS Pathog. 2010. PMID: 20976203 Free PMC article.
-
RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP.Nucleic Acids Res. 2013 Apr;41(8):4686-98. doi: 10.1093/nar/gkt159. Epub 2013 Mar 6. Nucleic Acids Res. 2013. PMID: 23471002 Free PMC article.
-
7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor.RNA Biol. 2009 Apr-Jun;6(2):122-8. doi: 10.4161/rna.6.2.8115. Epub 2009 Apr 6. RNA Biol. 2009. PMID: 19246988 Review.
-
The 7SK snRNP complex: a critical regulator in carcinogenesis.Biochimie. 2025 May 12:S0300-9084(25)00084-7. doi: 10.1016/j.biochi.2025.05.003. Online ahead of print. Biochimie. 2025. PMID: 40368082 Review.
Cited by
-
Chemical reversible crosslinking enables measurement of RNA 3D distances and alternative conformations in cells.Nat Commun. 2022 Feb 17;13(1):911. doi: 10.1038/s41467-022-28602-3. Nat Commun. 2022. PMID: 35177610 Free PMC article.
-
Reconstitution of a functional 7SK snRNP.Nucleic Acids Res. 2017 Jun 20;45(11):6864-6880. doi: 10.1093/nar/gkx262. Nucleic Acids Res. 2017. PMID: 28431135 Free PMC article.
-
RNA Modeling with the Computational Energy Landscape Framework.Methods Mol Biol. 2021;2323:49-66. doi: 10.1007/978-1-0716-1499-0_5. Methods Mol Biol. 2021. PMID: 34086273
-
Detailed secondary structure models of invertebrate 7SK RNAs.RNA Biol. 2018 Feb 1;15(2):158-164. doi: 10.1080/15476286.2017.1412913. Epub 2017 Dec 21. RNA Biol. 2018. PMID: 29219696 Free PMC article.
-
Computer-aided comprehensive explorations of RNA structural polymorphism through complementary simulation methods.QRB Discov. 2022 Oct 17;3:e21. doi: 10.1017/qrd.2022.19. eCollection 2022. QRB Discov. 2022. PMID: 37529277 Free PMC article. Review.
References
-
- Barboric M, Lenasi T. 2010. Kick-sTARting HIV-1 transcription elongation by 7SK snRNP deporTATion. Nat Struct Mol Biol 17: 928–930. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
