Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping
- PMID: 27853175
- PMCID: PMC5118543
- DOI: 10.1038/ncomms13342
Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping
Abstract
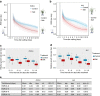
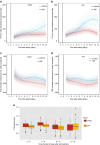
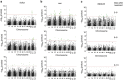
High-throughput phenotyping produces multiple measurements over time, which require new methods of analyses that are flexible in their quantification of plant growth and transpiration, yet are computationally economic. Here we develop such analyses and apply this to a rice population genotyped with a 700k SNP high-density array. Two rice diversity panels, indica and aus, containing a total of 553 genotypes, are phenotyped in waterlogged conditions. Using cubic smoothing splines to estimate plant growth and transpiration, we identify four time intervals that characterize the early responses of rice to salinity. Relative growth rate, transpiration rate and transpiration use efficiency (TUE) are analysed using a new association model that takes into account the interaction between treatment (control and salt) and genetic marker. This model allows the identification of previously undetected loci affecting TUE on chromosome 11, providing insights into the early responses of rice to salinity, in particular into the effects of salinity on plant growth and transpiration.
Figures
References
-
- Qadir M. et al. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 38, 282–295 (2014).
-
- Munns R. & Tester M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008). - PubMed
-
- Grattan S. R., Zeng L., Shannon M. C. & Roberts S. R. Rice is more sensitive to salinity than previously thought. Calif. Agric. 56, 189–195 (2002).
-
- Wicke B. et al. The global technical and economic potential of bioenergy from salt-affected soils. Energy Environ. Sci. 4, 2669 (2011).
-
- Vinod K. K., Krishnan S. G., Babu N. N., Nagarajan M. & Singh A. K. in Salt Stress in Plants eds Parvaiz Ahmad M., Azooz M., Prasad M. N. V. 219–260Springer (2013).
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

