Selective sweep on human amylase genes postdates the split with Neanderthals
- PMID: 27853181
- PMCID: PMC5112570
- DOI: 10.1038/srep37198
Selective sweep on human amylase genes postdates the split with Neanderthals
Abstract
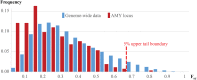
Humans have more copies of amylase genes than other primates. It is still poorly understood, however, when the copy number expansion occurred and whether its spread was enhanced by selection. Here we assess amylase copy numbers in a global sample of 480 high coverage genomes and find that regions flanking the amylase locus show notable depression of genetic diversity both in African and non-African populations. Analysis of genetic variation in these regions supports the model of an early selective sweep in the human lineage after the split of humans from Neanderthals which led to the fixation of multiple copies of AMY1 in place of a single copy. We find evidence of multiple secondary losses of copy number with the highest frequency (52%) of a deletion of AMY2A and associated low copy number of AMY1 in Northeast Siberian populations whose diet has been low in starch content.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

