Aβ42 oligomers modulate β-secretase through an XBP-1s-dependent pathway involving HRD1
- PMID: 27853315
- PMCID: PMC5112606
- DOI: 10.1038/srep37436
Aβ42 oligomers modulate β-secretase through an XBP-1s-dependent pathway involving HRD1
Abstract
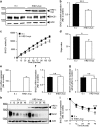
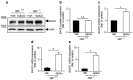
The aspartyl protease β-site APP cleaving enzyme, BACE1, is the rate-limiting enzyme involved in the production of amyloid-β peptide, which accumulates in both sporadic and familial cases of Alzheimer's disease and is at the center of gravity of the amyloid cascade hypothesis. In this context, unravelling the molecular mechanisms controlling BACE1 expression and activity in both physiological and pathological conditions remains of major importance. We previously demonstrated that Aβ controlled BACE1 transcription in an NFκB-dependent manner. Here, we delineate an additional cellular pathway by which natural and synthetic Aβ42 oligomers enhance active X-box binding protein XBP-1s. XBP-1s lowers BACE1 expression and activity indirectly, via the up-regulation of the ubiquitin-ligase HRD1 that acts as an endogenous down-regulator of BACE1. Thus, we delineate a novel pathway by which cells could compensate for Aβ42 oligomers production and thus, associated toxicity, by triggering a compensatory mechanism aimed at lowering BACE-1-mediated Aβ production by a molecular cascade involving XBP-1s and HRD1. It thus identifies HRD1 as a potential target for a novel Aβ-centered therapeutic strategy.
Figures

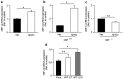

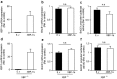
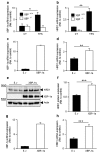
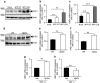
Similar articles
-
CHIP stabilizes amyloid precursor protein via proteasomal degradation and p53-mediated trans-repression of β-secretase.Aging Cell. 2015 Aug;14(4):595-604. doi: 10.1111/acel.12335. Epub 2015 Mar 13. Aging Cell. 2015. PMID: 25773675 Free PMC article.
-
Control of BACE1 degradation and APP processing by ubiquitin carboxyl-terminal hydrolase L1.J Neurochem. 2012 Mar;120(6):1129-38. doi: 10.1111/j.1471-4159.2011.07644.x. Epub 2012 Feb 10. J Neurochem. 2012. PMID: 22212137
-
Amyloid β-protein oligomers upregulate the β-secretase, BACE1, through a post-translational mechanism involving its altered subcellular distribution in neurons.Mol Brain. 2015 Nov 9;8(1):73. doi: 10.1186/s13041-015-0163-5. Mol Brain. 2015. PMID: 26552445 Free PMC article.
-
The beta-secretase, BACE: a prime drug target for Alzheimer's disease.J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review.
-
BACE1: the beta-secretase enzyme in Alzheimer's disease.J Mol Neurosci. 2004;23(1-2):105-14. doi: 10.1385/JMN:23:1-2:105. J Mol Neurosci. 2004. PMID: 15126696 Review.
Cited by
-
Insulin Resistance and Diabetes Mellitus in Alzheimer's Disease.Cells. 2021 May 18;10(5):1236. doi: 10.3390/cells10051236. Cells. 2021. PMID: 34069890 Free PMC article. Review.
-
Upregulation of the Sarco-Endoplasmic Reticulum Calcium ATPase 1 Truncated Isoform Plays a Pathogenic Role in Alzheimer's Disease.Cells. 2019 Nov 28;8(12):1539. doi: 10.3390/cells8121539. Cells. 2019. PMID: 31795302 Free PMC article.
-
Roles of Ubiquitin Ligases and Deubiquitylases in Alzheimer's Disease.Mol Neurobiol. 2025 Jun;62(6):7747-7761. doi: 10.1007/s12035-025-04739-2. Epub 2025 Feb 11. Mol Neurobiol. 2025. PMID: 39932514 Review.
-
Roles of XBP1s in Transcriptional Regulation of Target Genes.Biomedicines. 2021 Jul 8;9(7):791. doi: 10.3390/biomedicines9070791. Biomedicines. 2021. PMID: 34356855 Free PMC article. Review.
-
The unfolded protein response transcription factor XBP1s ameliorates Alzheimer's disease by improving synaptic function and proteostasis.Mol Ther. 2023 Jul 5;31(7):2240-2256. doi: 10.1016/j.ymthe.2023.03.028. Epub 2023 Apr 4. Mol Ther. 2023. PMID: 37016577 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

